Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
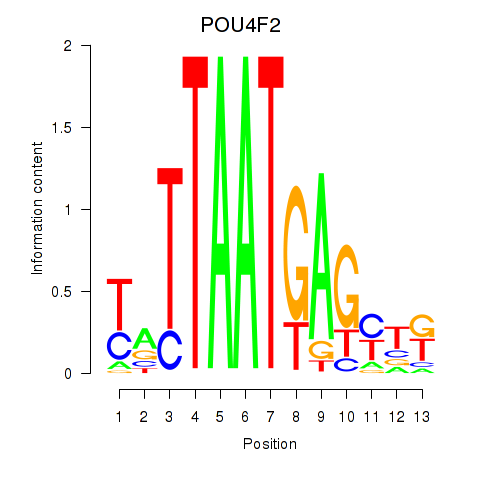
Results for POU4F2
Z-value: 0.93
Transcription factors associated with POU4F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F2
|
ENSG00000151615.3 | POU class 4 homeobox 2 |
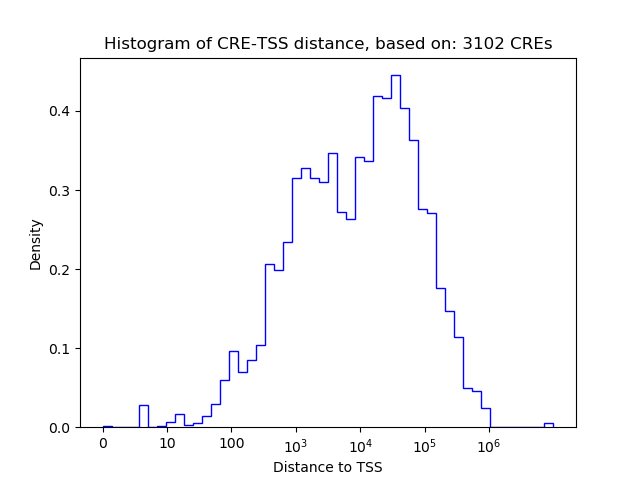
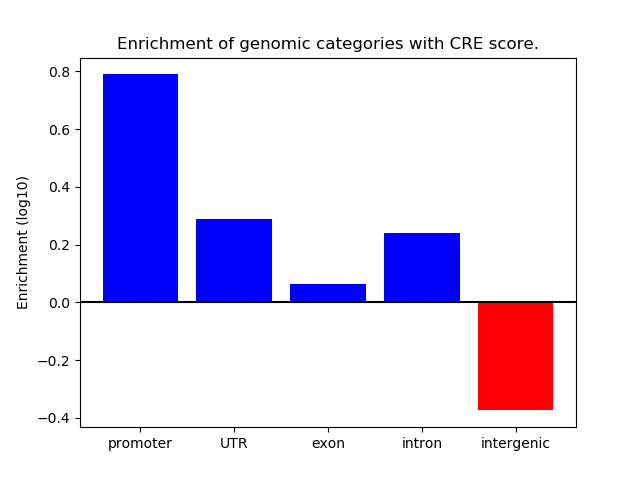
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_147559572_147559723 | POU4F2 | 398 | 0.699361 | 0.57 | 1.1e-01 | Click! |
| chr4_147559365_147559516 | POU4F2 | 605 | 0.600209 | 0.39 | 3.0e-01 | Click! |
Activity of the POU4F2 motif across conditions
Conditions sorted by the z-value of the POU4F2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
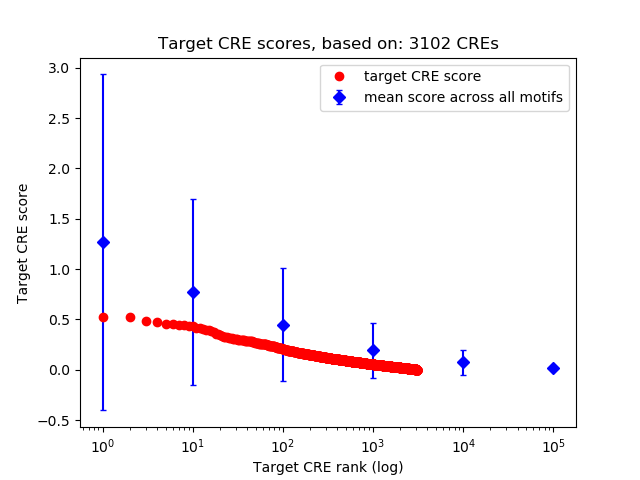
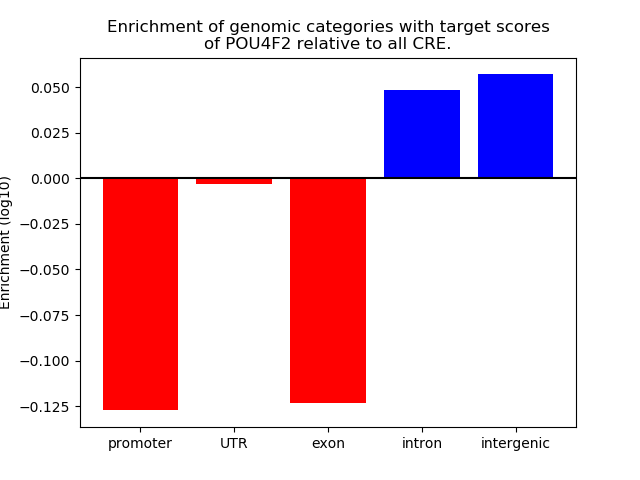
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_130587727_130588114 | 0.53 |
ENSG00000226380 |
. |
25622 |
0.22 |
| chr12_86229562_86230264 | 0.52 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
435 |
0.89 |
| chr18_55889704_55889927 | 0.48 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
1012 |
0.61 |
| chr8_55294582_55295235 | 0.47 |
ENSG00000244107 |
. |
38120 |
0.17 |
| chr14_29243474_29243665 | 0.46 |
C14orf23 |
chromosome 14 open reading frame 23 |
1568 |
0.33 |
| chr20_14315841_14316127 | 0.45 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
2270 |
0.36 |
| chr18_8707466_8707765 | 0.45 |
RP11-674N23.1 |
|
4 |
0.73 |
| chr3_98700853_98701271 | 0.45 |
ENSG00000207331 |
. |
73759 |
0.1 |
| chr1_48484300_48484451 | 0.44 |
TRABD2B |
TraB domain containing 2B |
21808 |
0.25 |
| chr21_28215844_28216573 | 0.43 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
112 |
0.98 |
| chr8_37350646_37351319 | 0.41 |
RP11-150O12.6 |
|
23557 |
0.24 |
| chr16_27590300_27590451 | 0.41 |
KIAA0556 |
KIAA0556 |
5147 |
0.22 |
| chr4_77611462_77611724 | 0.41 |
AC107072.2 |
|
52817 |
0.12 |
| chr3_98624953_98625332 | 0.40 |
ENSG00000207331 |
. |
2161 |
0.28 |
| chr12_3226358_3226555 | 0.39 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
32507 |
0.17 |
| chr15_49716998_49717158 | 0.38 |
FGF7 |
fibroblast growth factor 7 |
1621 |
0.44 |
| chr9_22239467_22239618 | 0.38 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
125865 |
0.06 |
| chr3_105657205_105657471 | 0.36 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
68942 |
0.15 |
| chr5_169398359_169398694 | 0.36 |
FAM196B |
family with sequence similarity 196, member B |
9218 |
0.26 |
| chr18_7572556_7572707 | 0.35 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
4814 |
0.33 |
| chr15_99297407_99297593 | 0.34 |
ENSG00000264480 |
. |
30155 |
0.19 |
| chr4_168818614_168818765 | 0.33 |
ANXA10 |
annexin A10 |
194977 |
0.03 |
| chr2_151335102_151335779 | 0.33 |
RND3 |
Rho family GTPase 3 |
6456 |
0.35 |
| chr7_1443631_1443991 | 0.33 |
MICALL2 |
MICAL-like 2 |
55151 |
0.11 |
| chr2_203882992_203883167 | 0.32 |
NBEAL1 |
neurobeachin-like 1 |
3477 |
0.29 |
| chr14_39961289_39961440 | 0.32 |
FBXO33 |
F-box protein 33 |
59660 |
0.14 |
| chr10_33622034_33622426 | 0.32 |
NRP1 |
neuropilin 1 |
1080 |
0.63 |
| chr20_48760619_48760901 | 0.31 |
TMEM189-UBE2V1 |
TMEM189-UBE2V1 readthrough |
9414 |
0.17 |
| chr10_14730521_14730672 | 0.31 |
ENSG00000201766 |
. |
25103 |
0.19 |
| chrX_135230667_135231011 | 0.31 |
FHL1 |
four and a half LIM domains 1 |
102 |
0.97 |
| chr7_134330319_134330562 | 0.30 |
BPGM |
2,3-bisphosphoglycerate mutase |
1120 |
0.61 |
| chr2_216484448_216484719 | 0.30 |
AC012462.1 |
|
183607 |
0.03 |
| chr11_106239153_106239583 | 0.30 |
RP11-680E19.1 |
|
104326 |
0.08 |
| chr3_75955654_75955867 | 0.30 |
ROBO2 |
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
86 |
0.98 |
| chr5_143138706_143138857 | 0.30 |
HMHB1 |
histocompatibility (minor) HB-1 |
52945 |
0.16 |
| chr5_141701461_141701673 | 0.29 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
2180 |
0.34 |
| chr8_66629390_66629680 | 0.29 |
RP11-707M3.3 |
|
2522 |
0.31 |
| chr16_64425240_64425489 | 0.29 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
668217 |
0.0 |
| chr9_21973547_21973827 | 0.29 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
1170 |
0.41 |
| chr8_41165312_41165463 | 0.29 |
SFRP1 |
secreted frizzled-related protein 1 |
878 |
0.57 |
| chr10_6763782_6764322 | 0.29 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr6_170596064_170596215 | 0.29 |
DLL1 |
delta-like 1 (Drosophila) |
3422 |
0.21 |
| chr12_26266071_26266589 | 0.29 |
SSPN |
sarcospan |
8594 |
0.18 |
| chr3_41244711_41244955 | 0.29 |
CTNNB1 |
catenin (cadherin-associated protein), beta 1, 88kDa |
3231 |
0.41 |
| chr11_2834821_2835116 | 0.28 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
47830 |
0.11 |
| chr6_160457314_160457599 | 0.28 |
AIRN |
antisense of IGF2R non-protein coding RNA |
28760 |
0.19 |
| chr1_24621564_24621715 | 0.27 |
ENSG00000266511 |
. |
15294 |
0.15 |
| chr12_52301279_52301690 | 0.27 |
ACVRL1 |
activin A receptor type II-like 1 |
282 |
0.9 |
| chr7_17720120_17720505 | 0.27 |
SNX13 |
sorting nexin 13 |
259779 |
0.02 |
| chr6_28442401_28443147 | 0.27 |
ZSCAN23 |
zinc finger and SCAN domain containing 23 |
31530 |
0.13 |
| chr6_69158334_69158485 | 0.27 |
ENSG00000266675 |
. |
44922 |
0.21 |
| chr7_42145445_42145596 | 0.27 |
GLI3 |
GLI family zinc finger 3 |
121800 |
0.07 |
| chr1_116472827_116472978 | 0.26 |
SLC22A15 |
solute carrier family 22, member 15 |
46217 |
0.16 |
| chr9_9701186_9701337 | 0.26 |
ENSG00000265735 |
. |
259201 |
0.02 |
| chr21_29050610_29050761 | 0.26 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
711853 |
0.0 |
| chr3_73085691_73085907 | 0.26 |
ENSG00000238959 |
. |
8840 |
0.16 |
| chr5_134914483_134914805 | 0.26 |
CXCL14 |
chemokine (C-X-C motif) ligand 14 |
110 |
0.67 |
| chr16_82049841_82049992 | 0.26 |
SDR42E1 |
short chain dehydrogenase/reductase family 42E, member 1 |
4823 |
0.21 |
| chr5_78808053_78808300 | 0.26 |
HOMER1 |
homer homolog 1 (Drosophila) |
441 |
0.86 |
| chr17_15301686_15301923 | 0.26 |
ENSG00000239888 |
. |
9897 |
0.2 |
| chr12_47097195_47097346 | 0.26 |
SLC38A4 |
solute carrier family 38, member 4 |
65817 |
0.15 |
| chr15_33130691_33130971 | 0.25 |
FMN1 |
formin 1 |
49624 |
0.13 |
| chr6_7544134_7544343 | 0.25 |
DSP |
desmoplakin |
2388 |
0.28 |
| chr4_181928076_181928259 | 0.25 |
ENSG00000251742 |
. |
828094 |
0.0 |
| chr9_22239273_22239447 | 0.25 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
125683 |
0.06 |
| chr17_42214986_42215221 | 0.25 |
ENSG00000212446 |
. |
1746 |
0.18 |
| chr8_57122510_57122859 | 0.25 |
PLAG1 |
pleiomorphic adenoma gene 1 |
1154 |
0.41 |
| chr15_59663421_59663716 | 0.25 |
FAM81A |
family with sequence similarity 81, member A |
1324 |
0.28 |
| chr10_60275835_60276146 | 0.24 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
3090 |
0.38 |
| chr11_131780111_131780399 | 0.24 |
NTM |
neurotrimin |
642 |
0.77 |
| chr9_108212044_108212195 | 0.24 |
FSD1L |
fibronectin type III and SPRY domain containing 1-like |
1715 |
0.48 |
| chr4_78093910_78094061 | 0.24 |
CCNG2 |
cyclin G2 |
14406 |
0.22 |
| chr21_36749606_36749887 | 0.24 |
RUNX1 |
runt-related transcription factor 1 |
328105 |
0.01 |
| chr1_68214173_68214559 | 0.24 |
ENSG00000238778 |
. |
23970 |
0.19 |
| chr13_27936373_27936791 | 0.24 |
ENSG00000252247 |
. |
18699 |
0.15 |
| chr10_116443956_116444184 | 0.23 |
ABLIM1 |
actin binding LIM protein 1 |
344 |
0.91 |
| chr11_85436820_85436971 | 0.23 |
SYTL2 |
synaptotagmin-like 2 |
616 |
0.73 |
| chr4_157691847_157691998 | 0.23 |
RP11-154F14.2 |
|
70589 |
0.11 |
| chr9_4137636_4137913 | 0.23 |
GLIS3 |
GLIS family zinc finger 3 |
7419 |
0.28 |
| chr1_100111779_100111966 | 0.23 |
PALMD |
palmdelphin |
123 |
0.98 |
| chr10_68587107_68587258 | 0.23 |
LRRTM3 |
leucine rich repeat transmembrane neuronal 3 |
98582 |
0.09 |
| chr7_70201396_70202343 | 0.23 |
AUTS2 |
autism susceptibility candidate 2 |
7744 |
0.34 |
| chrX_114859841_114860486 | 0.23 |
PLS3 |
plastin 3 |
13867 |
0.21 |
| chr9_18472965_18473116 | 0.22 |
ADAMTSL1 |
ADAMTS-like 1 |
852 |
0.75 |
| chr1_85247835_85247986 | 0.22 |
ENSG00000251899 |
. |
10972 |
0.25 |
| chr3_112931196_112931871 | 0.22 |
BOC |
BOC cell adhesion associated, oncogene regulated |
124 |
0.97 |
| chr13_68202936_68203087 | 0.22 |
PCDH9 |
protocadherin 9 |
398543 |
0.01 |
| chr14_55569037_55569677 | 0.22 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
21471 |
0.19 |
| chr5_21818159_21818310 | 0.22 |
ENSG00000221615 |
. |
83914 |
0.11 |
| chr4_149087033_149087184 | 0.22 |
RP11-76G10.1 |
|
19486 |
0.29 |
| chr6_138430146_138430297 | 0.22 |
PERP |
PERP, TP53 apoptosis effector |
1573 |
0.49 |
| chr16_86599945_86600151 | 0.22 |
FOXC2 |
forkhead box C2 (MFH-1, mesenchyme forkhead 1) |
809 |
0.48 |
| chr3_170807489_170807640 | 0.22 |
ENSG00000207963 |
. |
16984 |
0.23 |
| chr3_58983787_58983938 | 0.21 |
C3orf67 |
chromosome 3 open reading frame 67 |
51855 |
0.18 |
| chr3_155570793_155570970 | 0.21 |
SLC33A1 |
solute carrier family 33 (acetyl-CoA transporter), member 1 |
399 |
0.87 |
| chr11_9587935_9588242 | 0.21 |
WEE1 |
WEE1 G2 checkpoint kinase |
7140 |
0.16 |
| chr12_18841597_18841748 | 0.21 |
PLCZ1 |
phospholipase C, zeta 1 |
6299 |
0.25 |
| chr6_140065137_140065288 | 0.21 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
369455 |
0.01 |
| chr2_163171316_163171467 | 0.21 |
IFIH1 |
interferon induced with helicase C domain 1 |
3803 |
0.25 |
| chr4_89532288_89532605 | 0.21 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
5501 |
0.2 |
| chr21_17909068_17909746 | 0.21 |
ENSG00000207638 |
. |
2002 |
0.33 |
| chr1_229506200_229506351 | 0.20 |
ENSG00000252506 |
. |
12600 |
0.14 |
| chr8_18870397_18870669 | 0.20 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
663 |
0.74 |
| chr15_96886136_96886425 | 0.20 |
ENSG00000222651 |
. |
9790 |
0.16 |
| chr2_184434080_184434231 | 0.20 |
ENSG00000238306 |
. |
380008 |
0.01 |
| chr21_34918443_34918594 | 0.20 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
2721 |
0.18 |
| chr13_52218829_52218980 | 0.20 |
ENSG00000242893 |
. |
50553 |
0.11 |
| chr15_101668711_101668940 | 0.20 |
RP11-505E24.2 |
|
42554 |
0.16 |
| chr22_45406041_45406799 | 0.20 |
PHF21B |
PHD finger protein 21B |
540 |
0.81 |
| chr11_127461004_127461155 | 0.19 |
ENSG00000223315 |
. |
183333 |
0.03 |
| chr2_46561817_46561968 | 0.19 |
EPAS1 |
endothelial PAS domain protein 1 |
37351 |
0.19 |
| chr10_77648848_77649145 | 0.19 |
ENSG00000215921 |
. |
65460 |
0.13 |
| chr7_28355289_28355446 | 0.19 |
CREB5 |
cAMP responsive element binding protein 5 |
16427 |
0.26 |
| chr2_39751073_39751342 | 0.19 |
AC007246.3 |
|
5391 |
0.29 |
| chr17_58821066_58821723 | 0.19 |
ENSG00000252534 |
. |
5013 |
0.22 |
| chr3_112803505_112803695 | 0.19 |
ENSG00000238593 |
. |
7772 |
0.2 |
| chr17_65240369_65240671 | 0.19 |
RP11-401F2.3 |
|
433 |
0.62 |
| chr1_164532131_164532342 | 0.19 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
194 |
0.97 |
| chr6_113734377_113734528 | 0.19 |
ENSG00000222677 |
. |
101737 |
0.08 |
| chr10_69831641_69832005 | 0.19 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
2071 |
0.33 |
| chr7_73246766_73246917 | 0.19 |
CLDN4 |
claudin 4 |
1648 |
0.31 |
| chr6_108486676_108487297 | 0.19 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
58 |
0.78 |
| chr12_56917781_56918152 | 0.19 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
2183 |
0.23 |
| chr4_183370400_183371073 | 0.19 |
TENM3 |
teneurin transmembrane protein 3 |
584 |
0.84 |
| chr7_32929941_32930760 | 0.19 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
415 |
0.87 |
| chr8_59027426_59027596 | 0.18 |
FAM110B |
family with sequence similarity 110, member B |
120398 |
0.07 |
| chr2_52845575_52845726 | 0.18 |
ENSG00000264975 |
. |
84103 |
0.11 |
| chr1_70672840_70672991 | 0.18 |
SRSF11 |
serine/arginine-rich splicing factor 11 |
1497 |
0.3 |
| chr2_163157606_163157781 | 0.18 |
IFIH1 |
interferon induced with helicase C domain 1 |
17501 |
0.19 |
| chr5_135470545_135470952 | 0.18 |
SMAD5-AS1 |
SMAD5 antisense RNA 1 |
169 |
0.92 |
| chr3_189509857_189510008 | 0.18 |
TP63 |
tumor protein p63 |
2342 |
0.39 |
| chr7_17701857_17702150 | 0.18 |
SNX13 |
sorting nexin 13 |
278088 |
0.01 |
| chr7_70048822_70048973 | 0.18 |
AUTS2 |
autism susceptibility candidate 2 |
145228 |
0.05 |
| chr21_24278413_24278564 | 0.18 |
ENSG00000263796 |
. |
225496 |
0.02 |
| chr2_200446222_200446758 | 0.18 |
SATB2 |
SATB homeobox 2 |
110501 |
0.07 |
| chr7_150785302_150785601 | 0.18 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
1297 |
0.25 |
| chr8_94931050_94931201 | 0.18 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
13 |
0.98 |
| chr13_24782412_24782563 | 0.18 |
SPATA13 |
spermatogenesis associated 13 |
43345 |
0.13 |
| chr4_14728626_14728777 | 0.18 |
ENSG00000202449 |
. |
36367 |
0.24 |
| chr14_27067998_27068288 | 0.18 |
NOVA1-AS1 |
NOVA1 antisense RNA 1 (head to head) |
525 |
0.68 |
| chr10_79265125_79265301 | 0.18 |
ENSG00000199592 |
. |
81594 |
0.1 |
| chr11_103677106_103677422 | 0.17 |
ENSG00000264200 |
. |
43370 |
0.2 |
| chr3_159484310_159484461 | 0.17 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
1493 |
0.34 |
| chr6_16737749_16737900 | 0.17 |
RP1-151F17.1 |
|
23545 |
0.22 |
| chr5_87780535_87780686 | 0.17 |
TMEM161B-AS1 |
TMEM161B antisense RNA 1 |
58098 |
0.14 |
| chr16_54968513_54968925 | 0.17 |
IRX5 |
iroquois homeobox 5 |
2735 |
0.43 |
| chr8_10568187_10568338 | 0.17 |
CTD-2135J3.3 |
|
18562 |
0.12 |
| chr12_104613330_104613526 | 0.17 |
TXNRD1 |
thioredoxin reductase 1 |
3869 |
0.26 |
| chr3_124421533_124421684 | 0.17 |
ENSG00000263775 |
. |
5725 |
0.16 |
| chr2_37798390_37798541 | 0.17 |
AC006369.2 |
|
28814 |
0.21 |
| chr4_148657265_148657705 | 0.17 |
ARHGAP10 |
Rho GTPase activating protein 10 |
4271 |
0.21 |
| chr3_159483781_159484285 | 0.17 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
1141 |
0.44 |
| chr10_16992831_16993197 | 0.17 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
33180 |
0.22 |
| chr15_33154166_33154394 | 0.17 |
FMN1 |
formin 1 |
26175 |
0.19 |
| chr3_185446261_185446412 | 0.17 |
C3orf65 |
chromosome 3 open reading frame 65 |
15256 |
0.21 |
| chr9_111249319_111249470 | 0.17 |
ENSG00000222512 |
. |
128185 |
0.06 |
| chr22_36232810_36232961 | 0.17 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3380 |
0.35 |
| chr7_80542898_80543049 | 0.17 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
5526 |
0.35 |
| chr5_79542586_79542846 | 0.17 |
ENSG00000239159 |
. |
7308 |
0.19 |
| chr5_109671354_109671505 | 0.17 |
ENSG00000221436 |
. |
178187 |
0.03 |
| chr1_149378983_149379172 | 0.16 |
FCGR1C |
Fc fragment of IgG, high affinity Ic, receptor (CD64), pseudogene |
9720 |
0.13 |
| chr20_19966352_19966503 | 0.16 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
31333 |
0.18 |
| chr18_66505598_66505749 | 0.16 |
CCDC102B |
coiled-coil domain containing 102B |
1672 |
0.41 |
| chr2_233453880_233454031 | 0.16 |
AC073254.1 |
|
2577 |
0.17 |
| chr14_88738980_88739131 | 0.16 |
KCNK10 |
potassium channel, subfamily K, member 10 |
1800 |
0.48 |
| chr5_92918170_92918321 | 0.16 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
798 |
0.54 |
| chr19_56332953_56333104 | 0.16 |
NLRP11 |
NLR family, pyrin domain containing 11 |
3425 |
0.16 |
| chrX_107680802_107681081 | 0.16 |
COL4A6 |
collagen, type IV, alpha 6 |
615 |
0.74 |
| chr15_101069679_101069830 | 0.16 |
CERS3 |
ceramide synthase 3 |
14722 |
0.15 |
| chr11_12696722_12697409 | 0.16 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
15 |
0.99 |
| chr1_68686585_68686736 | 0.16 |
WLS |
wntless Wnt ligand secretion mediator |
10710 |
0.23 |
| chr18_59665868_59666178 | 0.16 |
RNF152 |
ring finger protein 152 |
104559 |
0.07 |
| chr1_203648008_203648159 | 0.16 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
3854 |
0.27 |
| chr17_35851593_35851869 | 0.16 |
DUSP14 |
dual specificity phosphatase 14 |
161 |
0.96 |
| chr1_191327786_191327937 | 0.16 |
RGS18 |
regulator of G-protein signaling 18 |
799726 |
0.0 |
| chr6_114181963_114182196 | 0.16 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
3538 |
0.24 |
| chr6_125420988_125421372 | 0.16 |
TPD52L1 |
tumor protein D52-like 1 |
19015 |
0.24 |
| chr10_5005306_5005493 | 0.16 |
AKR1C1 |
aldo-keto reductase family 1, member C1 |
46 |
0.92 |
| chr3_58038318_58038519 | 0.16 |
FLNB |
filamin B, beta |
25638 |
0.22 |
| chr2_205529494_205529645 | 0.16 |
PARD3B |
par-3 family cell polarity regulator beta |
118846 |
0.07 |
| chr1_178027513_178027706 | 0.16 |
RASAL2 |
RAS protein activator like 2 |
35255 |
0.22 |
| chr15_74880905_74881056 | 0.16 |
CLK3 |
CDC-like kinase 3 |
19733 |
0.14 |
| chr15_96897591_96898313 | 0.15 |
AC087477.1 |
Uncharacterized protein |
6535 |
0.18 |
| chr10_5045552_5046386 | 0.15 |
AKR1C2 |
aldo-keto reductase family 1, member C2 |
83 |
0.93 |
| chr17_69531749_69531900 | 0.15 |
ENSG00000222563 |
. |
224515 |
0.02 |
| chr9_71790192_71790670 | 0.15 |
TJP2 |
tight junction protein 2 |
1221 |
0.59 |
| chr17_63550102_63550306 | 0.15 |
AXIN2 |
axin 2 |
4643 |
0.32 |
| chr5_123149834_123150053 | 0.15 |
CSNK1G3 |
casein kinase 1, gamma 3 |
225824 |
0.02 |
| chr3_197121898_197122049 | 0.15 |
ENSG00000238491 |
. |
55551 |
0.12 |
| chr20_30309971_30310242 | 0.15 |
BCL2L1 |
BCL2-like 1 |
245 |
0.84 |
| chr4_152331570_152331721 | 0.15 |
FAM160A1 |
family with sequence similarity 160, member A1 |
1141 |
0.61 |
| chr18_53067170_53067321 | 0.15 |
TCF4 |
transcription factor 4 |
1321 |
0.53 |
| chr1_55183020_55183171 | 0.15 |
TTC4 |
tetratricopeptide repeat domain 4 |
1600 |
0.36 |
| chr3_98605225_98605614 | 0.15 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
14596 |
0.17 |
| chr4_44656889_44657040 | 0.15 |
YIPF7 |
Yip1 domain family, member 7 |
3306 |
0.24 |
| chr2_179144221_179144372 | 0.15 |
OSBPL6 |
oxysterol binding protein-like 6 |
5358 |
0.27 |
| chr1_220527416_220527567 | 0.15 |
AC096644.1 |
Uncharacterized protein |
80532 |
0.09 |
| chr10_74600201_74600352 | 0.15 |
OIT3 |
oncoprotein induced transcript 3 |
53063 |
0.12 |
| chr3_87036535_87037613 | 0.15 |
VGLL3 |
vestigial like 3 (Drosophila) |
2778 |
0.42 |
| chr5_124820548_124820891 | 0.15 |
ENSG00000222107 |
. |
133895 |
0.06 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.3 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.2 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.2 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.1 | 0.4 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.1 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0072141 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.0 | GO:0003160 | endocardium development(GO:0003157) endocardium morphogenesis(GO:0003160) endocardium formation(GO:0060214) |
| 0.0 | 0.1 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.2 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus(GO:0002866) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0010662 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.2 | GO:0048821 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.2 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) intestinal lipid absorption(GO:0098856) |
| 0.0 | 0.1 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0032589 | neuron projection membrane(GO:0032589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.3 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0051184 | cofactor transporter activity(GO:0051184) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |