Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
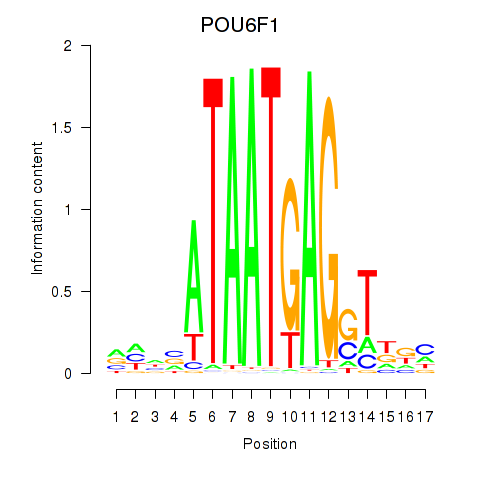
Results for POU6F1
Z-value: 1.32
Transcription factors associated with POU6F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F1
|
ENSG00000184271.11 | POU class 6 homeobox 1 |
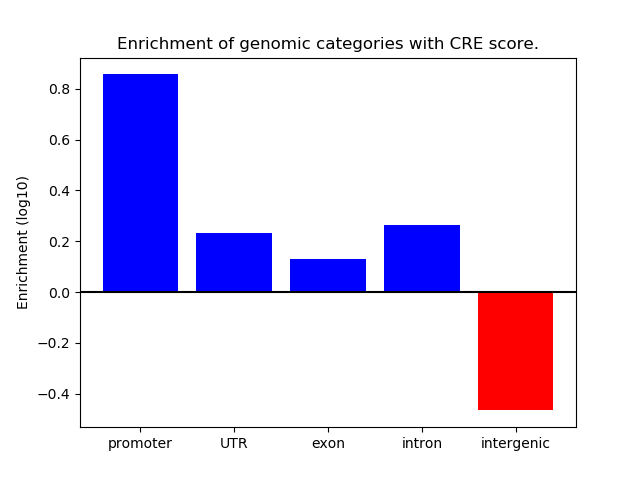
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_51611354_51611505 | POU6F1 | 48 | 0.962858 | -0.54 | 1.3e-01 | Click! |
| chr12_51611544_51611860 | POU6F1 | 225 | 0.898800 | -0.47 | 2.0e-01 | Click! |
| chr12_51610572_51610723 | POU6F1 | 830 | 0.510628 | -0.46 | 2.1e-01 | Click! |
| chr12_51610343_51610494 | POU6F1 | 1059 | 0.412881 | 0.37 | 3.3e-01 | Click! |
| chr12_51610880_51611170 | POU6F1 | 452 | 0.747794 | 0.14 | 7.1e-01 | Click! |
Activity of the POU6F1 motif across conditions
Conditions sorted by the z-value of the POU6F1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
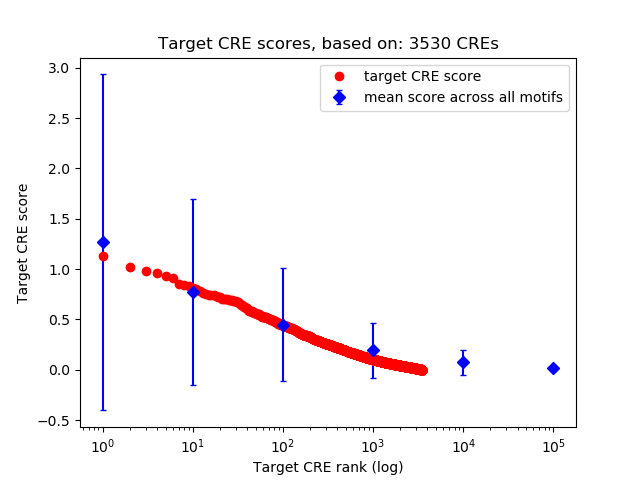
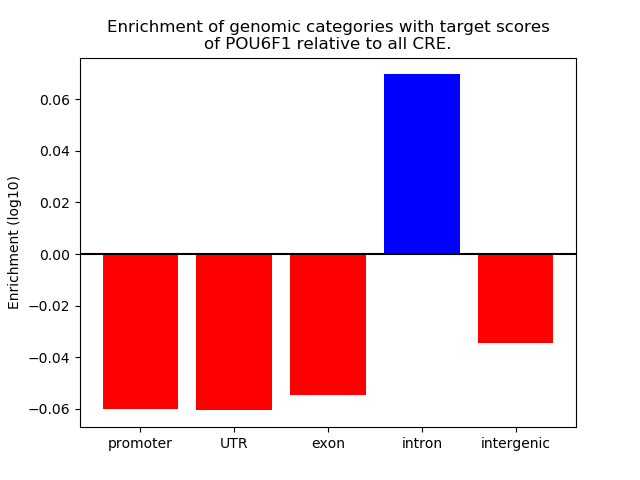
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_160766445_160766727 | 1.13 |
LY9 |
lymphocyte antigen 9 |
609 |
0.72 |
| chr6_90790442_90790643 | 1.03 |
ENSG00000222078 |
. |
79317 |
0.1 |
| chrX_11778976_11779311 | 0.98 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
1396 |
0.58 |
| chr19_6481304_6482171 | 0.96 |
DENND1C |
DENN/MADD domain containing 1C |
27 |
0.95 |
| chr5_169720489_169720914 | 0.93 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
4530 |
0.26 |
| chr2_233926827_233927087 | 0.91 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
1768 |
0.35 |
| chr1_100820231_100820573 | 0.85 |
CDC14A |
cell division cycle 14A |
1897 |
0.33 |
| chrX_78400540_78401325 | 0.85 |
GPR174 |
G protein-coupled receptor 174 |
25537 |
0.27 |
| chr4_40201423_40201705 | 0.83 |
RHOH |
ras homolog family member H |
400 |
0.87 |
| chrX_41779133_41779284 | 0.81 |
ENSG00000251807 |
. |
606 |
0.77 |
| chr12_54891959_54892179 | 0.80 |
NCKAP1L |
NCK-associated protein 1-like |
499 |
0.73 |
| chr12_9912315_9912520 | 0.79 |
CD69 |
CD69 molecule |
1080 |
0.47 |
| chr10_73529493_73529671 | 0.76 |
C10orf54 |
chromosome 10 open reading frame 54 |
3673 |
0.23 |
| chr5_130413183_130413334 | 0.75 |
HINT1 |
histidine triad nucleotide binding protein 1 |
85062 |
0.1 |
| chr14_22977314_22977663 | 0.74 |
TRAJ15 |
T cell receptor alpha joining 15 |
21092 |
0.09 |
| chrX_56590218_56591666 | 0.74 |
UBQLN2 |
ubiquilin 2 |
916 |
0.74 |
| chr2_106392161_106392312 | 0.74 |
NCK2 |
NCK adaptor protein 2 |
30048 |
0.23 |
| chr15_86235334_86235485 | 0.73 |
RP11-815J21.1 |
|
8931 |
0.12 |
| chr6_35271134_35271290 | 0.72 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
5583 |
0.2 |
| chr19_48747420_48747628 | 0.72 |
CARD8 |
caspase recruitment domain family, member 8 |
3204 |
0.16 |
| chr14_55799126_55799413 | 0.71 |
RP11-665C16.5 |
|
6950 |
0.25 |
| chr7_38386000_38386394 | 0.71 |
AMPH |
amphiphysin |
116516 |
0.06 |
| chr1_9477600_9477751 | 0.70 |
ENSG00000252956 |
. |
20162 |
0.21 |
| chr2_158299843_158300108 | 0.70 |
CYTIP |
cytohesin 1 interacting protein |
679 |
0.68 |
| chr2_136876136_136876323 | 0.69 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
494 |
0.87 |
| chr6_112078365_112078719 | 0.69 |
FYN |
FYN oncogene related to SRC, FGR, YES |
1775 |
0.47 |
| chr11_102192518_102192994 | 0.69 |
BIRC3 |
baculoviral IAP repeat containing 3 |
3196 |
0.22 |
| chr6_139458457_139458631 | 0.68 |
HECA |
headcase homolog (Drosophila) |
2295 |
0.39 |
| chr2_102973306_102973457 | 0.68 |
IL18R1 |
interleukin 18 receptor 1 |
992 |
0.54 |
| chr1_89664962_89665153 | 0.68 |
GBP4 |
guanylate binding protein 4 |
442 |
0.83 |
| chr6_119398487_119399078 | 0.67 |
FAM184A |
family with sequence similarity 184, member A |
755 |
0.67 |
| chr4_40209794_40209945 | 0.67 |
RHOH |
ras homolog family member H |
7905 |
0.22 |
| chr7_38381792_38381992 | 0.66 |
AMPH |
amphiphysin |
120821 |
0.06 |
| chr2_231193737_231193888 | 0.65 |
SP140L |
SP140 nuclear body protein-like |
1827 |
0.43 |
| chr20_39769733_39769905 | 0.64 |
RP1-1J6.2 |
|
3176 |
0.25 |
| chr7_38399403_38399654 | 0.63 |
AMPH |
amphiphysin |
103185 |
0.08 |
| chr7_142344805_142344957 | 0.63 |
MTRNR2L6 |
MT-RNR2-like 6 |
29223 |
0.21 |
| chr17_37933702_37934037 | 0.62 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
609 |
0.66 |
| chr5_39201497_39202174 | 0.62 |
FYB |
FYN binding protein |
1294 |
0.59 |
| chr10_30818195_30818576 | 0.61 |
ENSG00000239744 |
. |
26448 |
0.21 |
| chr2_182174133_182174359 | 0.60 |
ENSG00000266705 |
. |
3867 |
0.36 |
| chr1_160678018_160678169 | 0.58 |
CD48 |
CD48 molecule |
3500 |
0.2 |
| chr15_61013382_61013533 | 0.58 |
ENSG00000212625 |
. |
15511 |
0.2 |
| chr10_173487_173638 | 0.58 |
ZMYND11 |
zinc finger, MYND-type containing 11 |
6843 |
0.24 |
| chr3_63954232_63954647 | 0.58 |
ATXN7 |
ataxin 7 |
1019 |
0.48 |
| chr10_11208288_11208755 | 0.57 |
CELF2 |
CUGBP, Elav-like family member 2 |
1030 |
0.57 |
| chr15_34620156_34620427 | 0.57 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
8754 |
0.11 |
| chr17_80084198_80084349 | 0.57 |
CCDC57 |
coiled-coil domain containing 57 |
2159 |
0.16 |
| chr3_112692076_112692299 | 0.56 |
CD200R1 |
CD200 receptor 1 |
1572 |
0.33 |
| chr1_154303209_154303360 | 0.56 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
2009 |
0.18 |
| chr7_38393634_38393798 | 0.56 |
AMPH |
amphiphysin |
108997 |
0.07 |
| chr11_128343021_128343263 | 0.55 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
32147 |
0.21 |
| chr1_207509360_207509653 | 0.55 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
14370 |
0.25 |
| chr1_6087488_6087778 | 0.55 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
276 |
0.91 |
| chr12_739807_740337 | 0.55 |
RP11-218M22.1 |
|
15 |
0.97 |
| chr12_50908828_50909000 | 0.54 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
10024 |
0.23 |
| chr2_101558018_101558169 | 0.54 |
NPAS2 |
neuronal PAS domain protein 2 |
16469 |
0.19 |
| chr2_133917317_133917468 | 0.54 |
AC011755.1 |
HCG2006742; Protein LOC100996685 |
42815 |
0.19 |
| chr8_8727283_8727434 | 0.53 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
23797 |
0.18 |
| chr11_2322790_2323148 | 0.53 |
C11orf21 |
chromosome 11 open reading frame 21 |
174 |
0.62 |
| chr3_4536182_4536507 | 0.53 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
1110 |
0.44 |
| chr3_66444263_66444450 | 0.53 |
ENSG00000241587 |
. |
70077 |
0.12 |
| chr7_38343163_38343314 | 0.53 |
STARD3NL |
STARD3 N-terminal like |
125241 |
0.06 |
| chr3_114017077_114017228 | 0.52 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
3318 |
0.23 |
| chr14_99723235_99723386 | 0.52 |
AL109767.1 |
|
5975 |
0.23 |
| chr7_150146102_150146542 | 0.52 |
GIMAP8 |
GTPase, IMAP family member 8 |
1396 |
0.39 |
| chr6_114716123_114716274 | 0.52 |
RP3-399L15.3 |
|
30578 |
0.21 |
| chr16_28172650_28172801 | 0.52 |
XPO6 |
exportin 6 |
8497 |
0.18 |
| chr8_8655354_8655505 | 0.51 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
95726 |
0.07 |
| chr9_130735579_130735768 | 0.51 |
FAM102A |
family with sequence similarity 102, member A |
7119 |
0.11 |
| chr5_156617150_156617301 | 0.50 |
ITK |
IL2-inducible T-cell kinase |
9388 |
0.13 |
| chrY_7144223_7144374 | 0.50 |
PRKY |
protein kinase, Y-linked, pseudogene |
1944 |
0.4 |
| chr21_43640506_43640792 | 0.50 |
ENSG00000223262 |
. |
634 |
0.47 |
| chr7_144509813_144510191 | 0.50 |
TPK1 |
thiamin pyrophosphokinase 1 |
23144 |
0.23 |
| chr12_92528336_92528532 | 0.50 |
C12orf79 |
chromosome 12 open reading frame 79 |
2363 |
0.27 |
| chr14_61805647_61805798 | 0.49 |
PRKCH |
protein kinase C, eta |
5238 |
0.22 |
| chr3_108549304_108549455 | 0.49 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
7760 |
0.27 |
| chr4_150608043_150608194 | 0.49 |
ENSG00000202331 |
. |
220594 |
0.02 |
| chr2_219599622_219599773 | 0.49 |
TTLL4 |
tubulin tyrosine ligase-like family, member 4 |
2600 |
0.21 |
| chr7_130610833_130610984 | 0.49 |
ENSG00000226380 |
. |
48610 |
0.15 |
| chr1_150542961_150543112 | 0.48 |
ENSG00000264584 |
. |
1291 |
0.23 |
| chr20_54993887_54994066 | 0.48 |
CASS4 |
Cas scaffolding protein family member 4 |
6659 |
0.14 |
| chr7_38402576_38402839 | 0.48 |
AMPH |
amphiphysin |
100006 |
0.08 |
| chr14_22984428_22984696 | 0.48 |
TRAJ15 |
T cell receptor alpha joining 15 |
14018 |
0.1 |
| chr18_72662806_72662957 | 0.47 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
254587 |
0.02 |
| chr3_152002190_152002341 | 0.47 |
MBNL1-AS1 |
MBNL1 antisense RNA 1 |
14921 |
0.21 |
| chrX_78407976_78408214 | 0.46 |
GPR174 |
G protein-coupled receptor 174 |
18374 |
0.29 |
| chr18_2982488_2982849 | 0.46 |
LPIN2 |
lipin 2 |
203 |
0.93 |
| chr8_66843424_66843611 | 0.46 |
PDE7A |
phosphodiesterase 7A |
89330 |
0.09 |
| chr12_94805998_94806149 | 0.46 |
CCDC41 |
coiled-coil domain containing 41 |
47649 |
0.14 |
| chr5_4638073_4638224 | 0.46 |
ENSG00000223007 |
. |
488011 |
0.01 |
| chr13_48795647_48795882 | 0.46 |
ITM2B |
integral membrane protein 2B |
11530 |
0.26 |
| chr14_69821359_69821510 | 0.45 |
ERH |
enhancer of rudimentary homolog (Drosophila) |
43582 |
0.15 |
| chr22_20905629_20905785 | 0.45 |
MED15 |
mediator complex subunit 15 |
130 |
0.95 |
| chr8_13346130_13346281 | 0.45 |
DLC1 |
deleted in liver cancer 1 |
10545 |
0.24 |
| chr17_63225008_63225159 | 0.45 |
RGS9 |
regulator of G-protein signaling 9 |
91491 |
0.08 |
| chr6_109801994_109802145 | 0.45 |
ZBTB24 |
zinc finger and BTB domain containing 24 |
2371 |
0.18 |
| chr18_72873277_72873679 | 0.45 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
43990 |
0.18 |
| chr1_206732541_206732911 | 0.44 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
2233 |
0.27 |
| chr7_12601305_12601456 | 0.44 |
SCIN |
scinderin |
8932 |
0.23 |
| chr6_90791046_90791197 | 0.44 |
ENSG00000222078 |
. |
79896 |
0.1 |
| chr6_22249892_22250043 | 0.44 |
RP3-404K8.2 |
|
10686 |
0.27 |
| chr11_128589269_128589524 | 0.44 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23478 |
0.17 |
| chr7_151622563_151622783 | 0.44 |
ENSG00000222941 |
. |
9261 |
0.14 |
| chr1_158902534_158902685 | 0.43 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
1251 |
0.51 |
| chr9_100749769_100749920 | 0.43 |
RP11-535C21.3 |
|
94 |
0.96 |
| chr6_112076140_112076291 | 0.43 |
FYN |
FYN oncogene related to SRC, FGR, YES |
4102 |
0.32 |
| chr17_79258803_79259366 | 0.43 |
SLC38A10 |
solute carrier family 38, member 10 |
3020 |
0.17 |
| chr1_39025698_39025869 | 0.43 |
ENSG00000200796 |
. |
163452 |
0.04 |
| chr3_18483093_18483271 | 0.43 |
SATB1 |
SATB homeobox 1 |
2917 |
0.27 |
| chr6_45983691_45983842 | 0.43 |
CLIC5 |
chloride intracellular channel 5 |
180 |
0.95 |
| chr7_50267269_50267784 | 0.42 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
76798 |
0.1 |
| chr1_12445934_12446085 | 0.42 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
23915 |
0.22 |
| chr17_76767868_76768019 | 0.42 |
CYTH1 |
cytohesin 1 |
10411 |
0.19 |
| chr6_143867244_143867395 | 0.42 |
PHACTR2 |
phosphatase and actin regulator 2 |
9337 |
0.16 |
| chrX_106954029_106954352 | 0.42 |
TSC22D3 |
TSC22 domain family, member 3 |
5441 |
0.26 |
| chr10_54571770_54571921 | 0.42 |
MBL2 |
mannose-binding lectin (protein C) 2, soluble |
40385 |
0.17 |
| chr5_67577391_67577649 | 0.42 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
1385 |
0.58 |
| chr12_9908788_9909139 | 0.42 |
CD69 |
CD69 molecule |
4534 |
0.18 |
| chr12_14924776_14925101 | 0.42 |
HIST4H4 |
histone cluster 4, H4 |
873 |
0.43 |
| chr6_149805124_149805364 | 0.41 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
882 |
0.55 |
| chr9_77635535_77635794 | 0.41 |
RP11-197P3.5 |
|
4712 |
0.19 |
| chr6_45463753_45463944 | 0.41 |
RUNX2 |
runt-related transcription factor 2 |
73626 |
0.11 |
| chr10_111769664_111769909 | 0.41 |
ADD3 |
adducin 3 (gamma) |
2064 |
0.33 |
| chr17_35285773_35285924 | 0.41 |
RP11-445F12.1 |
|
8073 |
0.14 |
| chr3_37997723_37997874 | 0.41 |
CTDSPL |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
8272 |
0.16 |
| chr2_214004488_214004639 | 0.41 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
8790 |
0.3 |
| chrX_12969564_12969715 | 0.41 |
TMSB4X |
thymosin beta 4, X-linked |
23588 |
0.18 |
| chr4_36243817_36244102 | 0.41 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
1602 |
0.35 |
| chr3_56949638_56949992 | 0.41 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
684 |
0.75 |
| chr12_77165442_77165593 | 0.40 |
ZDHHC17 |
zinc finger, DHHC-type containing 17 |
7164 |
0.3 |
| chr6_27106968_27108062 | 0.40 |
HIST1H4I |
histone cluster 1, H4i |
439 |
0.71 |
| chr12_72523140_72523291 | 0.40 |
RP11-89M22.3 |
|
72058 |
0.12 |
| chrX_48773434_48773585 | 0.40 |
PIM2 |
pim-2 oncogene |
497 |
0.64 |
| chr2_238603556_238603753 | 0.39 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
2672 |
0.32 |
| chr2_44154306_44154579 | 0.39 |
LRPPRC |
leucine-rich pentatricopeptide repeat containing |
28019 |
0.17 |
| chr6_118985728_118985996 | 0.39 |
CEP85L |
centrosomal protein 85kDa-like |
12179 |
0.29 |
| chr12_19705618_19705769 | 0.39 |
AEBP2 |
AE binding protein 2 |
52630 |
0.17 |
| chr14_38558504_38558837 | 0.39 |
CTD-2058B24.2 |
|
1693 |
0.48 |
| chr9_125662893_125663901 | 0.39 |
RC3H2 |
ring finger and CCCH-type domains 2 |
3008 |
0.16 |
| chr2_75067589_75067763 | 0.39 |
HK2 |
hexokinase 2 |
5379 |
0.28 |
| chr1_100823462_100823613 | 0.39 |
CDC14A |
cell division cycle 14A |
5032 |
0.21 |
| chr7_133906611_133906762 | 0.38 |
AC008154.4 |
|
62566 |
0.13 |
| chr5_81074467_81075538 | 0.38 |
SSBP2 |
single-stranded DNA binding protein 2 |
27930 |
0.25 |
| chr11_118178183_118178334 | 0.38 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
2644 |
0.19 |
| chr3_111043169_111043480 | 0.38 |
CD96 |
CD96 molecule |
31758 |
0.24 |
| chr7_76822766_76822966 | 0.38 |
FGL2 |
fibrinogen-like 2 |
6277 |
0.24 |
| chr14_69281480_69282477 | 0.37 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
18788 |
0.19 |
| chr4_99525122_99525342 | 0.37 |
TSPAN5 |
tetraspanin 5 |
53554 |
0.14 |
| chr2_11859670_11859821 | 0.37 |
AC012456.3 |
|
4603 |
0.15 |
| chr5_20958214_20958365 | 0.37 |
CDH18 |
cadherin 18, type 2 |
382307 |
0.01 |
| chr11_3856061_3856212 | 0.37 |
RHOG |
ras homolog family member G |
2961 |
0.14 |
| chr9_101881575_101881851 | 0.37 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
8344 |
0.21 |
| chr6_139352039_139352190 | 0.37 |
ABRACL |
ABRA C-terminal like |
2135 |
0.38 |
| chr13_46752006_46752233 | 0.37 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
4340 |
0.19 |
| chr2_72438706_72438857 | 0.36 |
CYP26B1 |
cytochrome P450, family 26, subfamily B, polypeptide 1 |
63614 |
0.16 |
| chr11_34212282_34212433 | 0.36 |
ENSG00000201867 |
. |
1636 |
0.49 |
| chr9_100667248_100667478 | 0.36 |
C9orf156 |
chromosome 9 open reading frame 156 |
7390 |
0.17 |
| chr22_39238770_39238921 | 0.36 |
NPTXR |
neuronal pentraxin receptor |
1142 |
0.39 |
| chrX_78620829_78620980 | 0.36 |
ITM2A |
integral membrane protein 2A |
1952 |
0.51 |
| chr1_158902219_158902370 | 0.36 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
936 |
0.62 |
| chr4_90220151_90220996 | 0.35 |
GPRIN3 |
GPRIN family member 3 |
8588 |
0.31 |
| chr3_111261315_111261845 | 0.35 |
CD96 |
CD96 molecule |
583 |
0.82 |
| chr7_151048885_151049126 | 0.35 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
6574 |
0.16 |
| chr17_75178097_75178354 | 0.35 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
1106 |
0.55 |
| chr7_37382144_37382404 | 0.35 |
ELMO1 |
engulfment and cell motility 1 |
93 |
0.97 |
| chr14_24641513_24641917 | 0.35 |
REC8 |
REC8 meiotic recombination protein |
105 |
0.89 |
| chr3_114801119_114801270 | 0.35 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
10972 |
0.32 |
| chr13_24777878_24778029 | 0.35 |
ENSG00000252695 |
. |
41398 |
0.13 |
| chr14_39593348_39593499 | 0.35 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
9442 |
0.18 |
| chr13_41556565_41556847 | 0.35 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
288 |
0.92 |
| chr1_103470741_103470892 | 0.35 |
COL11A1 |
collagen, type XI, alpha 1 |
77599 |
0.13 |
| chr21_35885755_35885906 | 0.34 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
1257 |
0.46 |
| chr2_87813494_87813645 | 0.34 |
RP11-1399P15.1 |
|
36016 |
0.22 |
| chr5_100116536_100116687 | 0.34 |
ENSG00000221263 |
. |
35658 |
0.2 |
| chr18_56282753_56282991 | 0.34 |
ALPK2 |
alpha-kinase 2 |
13317 |
0.15 |
| chr1_215739347_215739498 | 0.34 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
1313 |
0.59 |
| chr1_237204411_237204562 | 0.34 |
RYR2 |
ryanodine receptor 2 (cardiac) |
1019 |
0.63 |
| chr1_169679248_169679951 | 0.34 |
SELL |
selectin L |
1240 |
0.48 |
| chr2_101944298_101944469 | 0.34 |
ENSG00000264857 |
. |
18471 |
0.16 |
| chr14_22993528_22993679 | 0.34 |
TRAJ15 |
T cell receptor alpha joining 15 |
4977 |
0.12 |
| chr7_65723704_65723855 | 0.34 |
TPST1 |
tyrosylprotein sulfotransferase 1 |
37188 |
0.16 |
| chrX_135738768_135738919 | 0.34 |
CD40LG |
CD40 ligand |
8457 |
0.18 |
| chrX_124428887_124429038 | 0.34 |
ENSG00000263886 |
. |
7337 |
0.34 |
| chr3_107844575_107844939 | 0.34 |
CD47 |
CD47 molecule |
34885 |
0.22 |
| chr1_209946189_209946340 | 0.34 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
4304 |
0.16 |
| chr7_56124875_56125026 | 0.34 |
ENSG00000206603 |
. |
1892 |
0.19 |
| chr7_130643743_130643906 | 0.34 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
25044 |
0.22 |
| chr8_131204031_131204182 | 0.34 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
10998 |
0.24 |
| chr9_74921921_74922072 | 0.34 |
ENSG00000252428 |
. |
7242 |
0.27 |
| chr12_12867727_12867916 | 0.34 |
CDKN1B |
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
2237 |
0.23 |
| chr12_105076421_105076798 | 0.33 |
ENSG00000264295 |
. |
91198 |
0.08 |
| chr2_220113848_220114166 | 0.33 |
STK16 |
serine/threonine kinase 16 |
3389 |
0.09 |
| chr5_130849992_130850161 | 0.33 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
6226 |
0.32 |
| chr14_22182172_22182323 | 0.33 |
OR4E2 |
olfactory receptor, family 4, subfamily E, member 2 |
48950 |
0.13 |
| chr3_74049848_74049999 | 0.33 |
ENSG00000212585 |
. |
165280 |
0.04 |
| chr10_3919669_3920010 | 0.33 |
KLF6 |
Kruppel-like factor 6 |
92366 |
0.09 |
| chr3_170984257_170984408 | 0.33 |
TNIK |
TRAF2 and NCK interacting kinase |
40832 |
0.2 |
| chr8_1920933_1921192 | 0.33 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
982 |
0.66 |
| chr10_70852755_70852906 | 0.33 |
SRGN |
serglycin |
4956 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.4 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.4 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.6 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.1 | 0.2 | GO:0014808 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.1 | 0.4 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.1 | 0.4 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.3 | GO:0051712 | positive regulation of killing of cells of other organism(GO:0051712) |
| 0.1 | 0.3 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.2 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.1 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 0.2 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.3 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.3 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 0.2 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.6 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.1 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 1.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.2 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 2.3 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0031342 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) negative regulation of cell killing(GO:0031342) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.1 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation involved in immune response(GO:0002313) mature B cell differentiation(GO:0002335) |
| 0.0 | 0.2 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.2 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.3 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.0 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0045606 | positive regulation of epidermal cell differentiation(GO:0045606) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 1.1 | GO:0016064 | immunoglobulin mediated immune response(GO:0016064) B cell mediated immunity(GO:0019724) |
| 0.0 | 0.0 | GO:0046885 | regulation of hormone biosynthetic process(GO:0046885) |
| 0.0 | 0.1 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0052306 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) positive regulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052555) positive regulation by symbiont of host immune response(GO:0052556) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0031442 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.1 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.3 | GO:0045762 | positive regulation of cyclase activity(GO:0031281) positive regulation of adenylate cyclase activity(GO:0045762) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0097502 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.1 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.0 | GO:0048537 | mucosal-associated lymphoid tissue development(GO:0048537) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0007140 | male meiosis(GO:0007140) |
| 0.0 | 0.0 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.0 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.4 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.2 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.0 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.0 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:1901998 | antibiotic transport(GO:0042891) toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0052200 | response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.3 | GO:0000800 | lateral element(GO:0000800) |
| 0.1 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.0 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.0 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.5 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.7 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.5 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.9 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.3 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.7 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.7 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.0 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.5 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.6 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0019209 | kinase activator activity(GO:0019209) |
| 0.0 | 0.0 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 1.2 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |