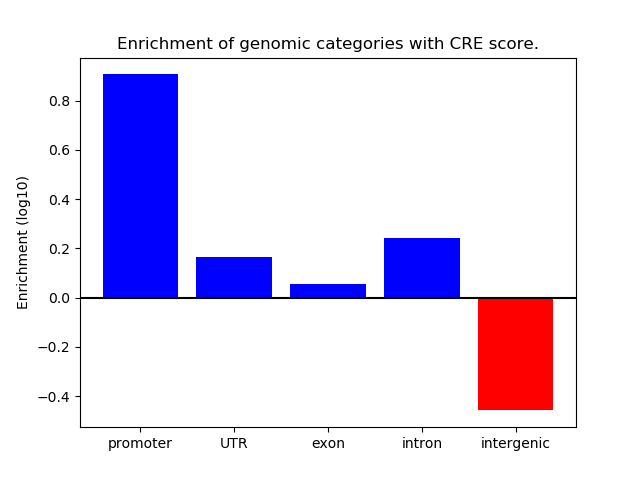
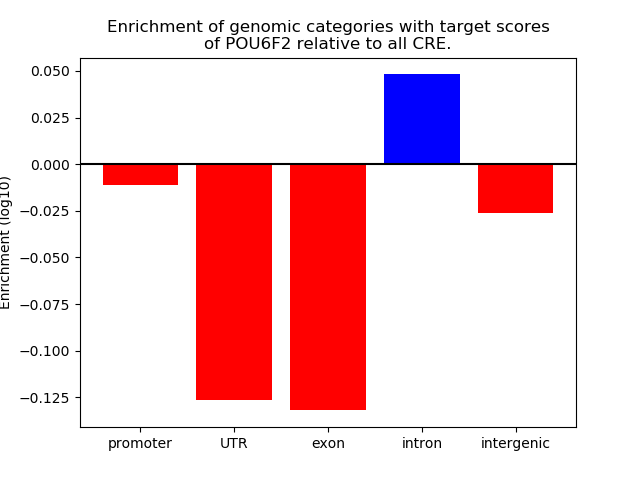
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
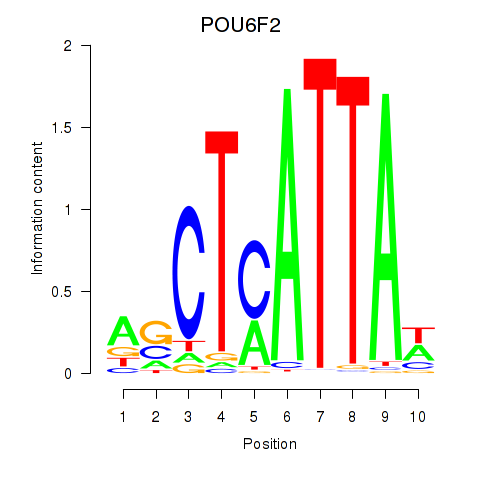
Results for POU6F2
Z-value: 0.27
Transcription factors associated with POU6F2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU6F2
|
ENSG00000106536.15 | POU class 6 homeobox 2 |
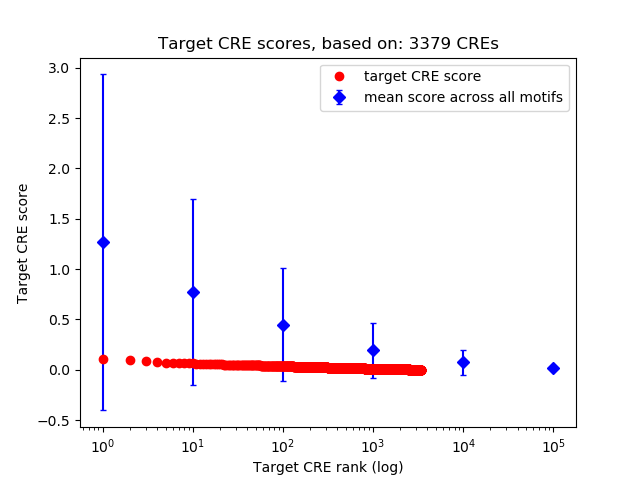
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_39125414_39125566 | POU6F2 | 29 | 0.986498 | 0.34 | 3.7e-01 | Click! |
| chr7_39170524_39170675 | POU6F2 | 45138 | 0.181268 | 0.31 | 4.2e-01 | Click! |
| chr7_39015718_39015994 | POU6F2 | 1742 | 0.421379 | 0.27 | 4.9e-01 | Click! |
| chr7_39170814_39170965 | POU6F2 | 45428 | 0.180424 | -0.11 | 7.7e-01 | Click! |
| chr7_39135393_39135544 | POU6F2 | 10007 | 0.280126 | -0.05 | 9.0e-01 | Click! |
Activity of the POU6F2 motif across conditions
Conditions sorted by the z-value of the POU6F2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
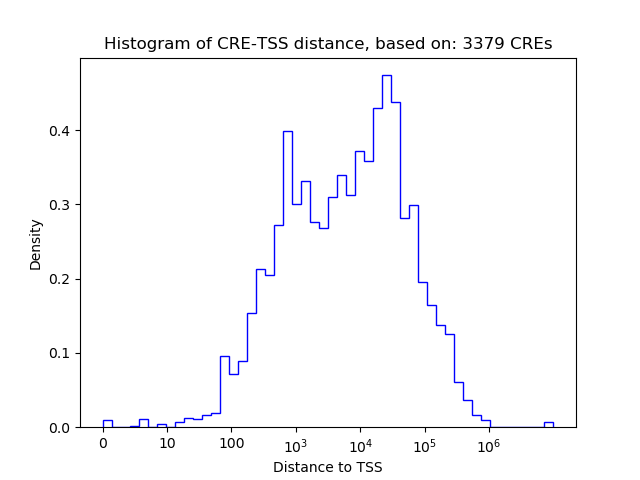
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_2653790_2653941 | 0.11 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
1872 |
0.28 |
| chr19_17907977_17908128 | 0.09 |
B3GNT3 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
2086 |
0.22 |
| chr3_160753561_160753761 | 0.09 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
65294 |
0.13 |
| chr9_23850343_23850766 | 0.07 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
24219 |
0.29 |
| chr11_98940627_98941137 | 0.07 |
CNTN5 |
contactin 5 |
48962 |
0.17 |
| chr5_124082726_124083211 | 0.07 |
ZNF608 |
zinc finger protein 608 |
648 |
0.62 |
| chr12_104613330_104613526 | 0.07 |
TXNRD1 |
thioredoxin reductase 1 |
3869 |
0.26 |
| chr12_12009100_12009251 | 0.07 |
ETV6 |
ets variant 6 |
29696 |
0.24 |
| chr6_36259798_36259949 | 0.07 |
PNPLA1 |
patatin-like phospholipase domain containing 1 |
21636 |
0.16 |
| chr1_225117686_225118141 | 0.06 |
DNAH14 |
dynein, axonemal, heavy chain 14 |
529 |
0.87 |
| chr4_57975072_57975270 | 0.06 |
IGFBP7-AS1 |
IGFBP7 antisense RNA 1 |
757 |
0.52 |
| chr13_27296169_27296320 | 0.06 |
GPR12 |
G protein-coupled receptor 12 |
38183 |
0.2 |
| chr11_8228040_8228278 | 0.06 |
RIC3 |
RIC3 acetylcholine receptor chaperone |
37557 |
0.16 |
| chr9_126781132_126781283 | 0.06 |
LHX2 |
LIM homeobox 2 |
3531 |
0.2 |
| chr4_77506894_77507309 | 0.06 |
ENSG00000265314 |
. |
10397 |
0.17 |
| chr1_198905629_198906537 | 0.06 |
ENSG00000207759 |
. |
77801 |
0.11 |
| chr9_90131536_90131777 | 0.06 |
DAPK1 |
death-associated protein kinase 1 |
17736 |
0.23 |
| chr8_56797598_56797816 | 0.05 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
5313 |
0.16 |
| chr1_85247835_85247986 | 0.05 |
ENSG00000251899 |
. |
10972 |
0.25 |
| chr2_17997369_17997520 | 0.05 |
MSGN1 |
mesogenin 1 |
319 |
0.9 |
| chr13_68202936_68203087 | 0.05 |
PCDH9 |
protocadherin 9 |
398543 |
0.01 |
| chr2_68618118_68618269 | 0.05 |
AC015969.3 |
|
25477 |
0.15 |
| chr3_104859509_104859660 | 0.05 |
ALCAM |
activated leukocyte cell adhesion molecule |
226169 |
0.02 |
| chr9_22239273_22239447 | 0.05 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
125683 |
0.06 |
| chr20_1892625_1892776 | 0.05 |
SIRPA |
signal-regulatory protein alpha |
16758 |
0.21 |
| chr9_90112157_90112308 | 0.05 |
DAPK1 |
death-associated protein kinase 1 |
89 |
0.98 |
| chr18_33057377_33057528 | 0.05 |
INO80C |
INO80 complex subunit C |
9982 |
0.21 |
| chr3_190057384_190057535 | 0.05 |
CLDN1 |
claudin 1 |
17195 |
0.23 |
| chr21_24278413_24278564 | 0.05 |
ENSG00000263796 |
. |
225496 |
0.02 |
| chr2_153362234_153362385 | 0.05 |
FMNL2 |
formin-like 2 |
113783 |
0.07 |
| chr2_169067281_169067721 | 0.05 |
STK39 |
serine threonine kinase 39 |
37150 |
0.22 |
| chr4_168818614_168818765 | 0.05 |
ANXA10 |
annexin A10 |
194977 |
0.03 |
| chr13_92002080_92002526 | 0.05 |
ENSG00000215417 |
. |
556 |
0.85 |
| chr3_155570793_155570970 | 0.05 |
SLC33A1 |
solute carrier family 33 (acetyl-CoA transporter), member 1 |
399 |
0.87 |
| chr2_60797281_60797571 | 0.05 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
16724 |
0.23 |
| chr3_171848594_171848745 | 0.05 |
FNDC3B |
fibronectin type III domain containing 3B |
3905 |
0.33 |
| chr11_66138436_66138603 | 0.05 |
SLC29A2 |
solute carrier family 29 (equilibrative nucleoside transporter), member 2 |
772 |
0.38 |
| chrX_9432067_9432218 | 0.05 |
TBL1X |
transducin (beta)-like 1X-linked |
771 |
0.79 |
| chr20_52271481_52272252 | 0.05 |
ENSG00000238468 |
. |
13431 |
0.22 |
| chr11_3856061_3856212 | 0.05 |
RHOG |
ras homolog family member G |
2961 |
0.14 |
| chr1_108594173_108594324 | 0.05 |
ENSG00000264753 |
. |
32855 |
0.19 |
| chr12_56692178_56692329 | 0.05 |
CS |
citrate synthase |
1505 |
0.16 |
| chr13_95752508_95752659 | 0.05 |
ENSG00000252335 |
. |
81094 |
0.1 |
| chr12_62604974_62605125 | 0.05 |
FAM19A2 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
18426 |
0.21 |
| chr14_75885513_75886422 | 0.05 |
RP11-293M10.6 |
|
8426 |
0.2 |
| chr11_33952138_33952289 | 0.05 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
38377 |
0.16 |
| chr6_150466476_150466683 | 0.05 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
2367 |
0.3 |
| chr20_56862949_56863100 | 0.05 |
PPP4R1L |
protein phosphatase 4, regulatory subunit 1-like |
21471 |
0.18 |
| chr18_60388254_60388405 | 0.05 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
5646 |
0.22 |
| chr3_159558086_159558308 | 0.04 |
SCHIP1 |
schwannomin interacting protein 1 |
547 |
0.76 |
| chrX_30646235_30646386 | 0.04 |
GK |
glycerol kinase |
25166 |
0.17 |
| chr11_118079199_118079350 | 0.04 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
4326 |
0.17 |
| chr2_33246915_33247066 | 0.04 |
ENSG00000252502 |
. |
30599 |
0.21 |
| chr20_43810439_43810590 | 0.04 |
PI3 |
peptidase inhibitor 3, skin-derived |
6997 |
0.15 |
| chr19_52253866_52254017 | 0.04 |
FPR1 |
formyl peptide receptor 1 |
123 |
0.92 |
| chr21_34886721_34886872 | 0.04 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
8615 |
0.12 |
| chr1_151432125_151432340 | 0.04 |
POGZ |
pogo transposable element with ZNF domain |
291 |
0.85 |
| chr5_37370451_37371178 | 0.04 |
NUP155 |
nucleoporin 155kDa |
73 |
0.98 |
| chr5_135170799_135170980 | 0.04 |
SLC25A48 |
solute carrier family 25, member 48 |
473 |
0.79 |
| chr12_42862422_42862573 | 0.04 |
RP11-328C8.4 |
|
9329 |
0.15 |
| chr4_38531163_38531332 | 0.04 |
RP11-617D20.1 |
|
94949 |
0.08 |
| chrX_112573298_112573449 | 0.04 |
ENSG00000238811 |
. |
329017 |
0.01 |
| chr6_49518031_49518578 | 0.04 |
C6orf141 |
chromosome 6 open reading frame 141 |
191 |
0.95 |
| chr18_37421688_37422106 | 0.04 |
ENSG00000212354 |
. |
163311 |
0.04 |
| chr18_55889704_55889927 | 0.04 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
1012 |
0.61 |
| chr14_35836928_35837079 | 0.04 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
36343 |
0.17 |
| chr13_61987939_61988245 | 0.04 |
PCDH20 |
protocadherin 20 |
1563 |
0.57 |
| chr3_196007930_196008081 | 0.04 |
PCYT1A |
phosphate cytidylyltransferase 1, choline, alpha |
6129 |
0.13 |
| chr6_136888235_136888386 | 0.04 |
MAP7 |
microtubule-associated protein 7 |
16353 |
0.15 |
| chr4_79473579_79473973 | 0.04 |
ANXA3 |
annexin A3 |
655 |
0.79 |
| chr22_20226360_20226511 | 0.04 |
RTN4R |
reticulon 4 receptor |
4772 |
0.15 |
| chr4_140765044_140765195 | 0.04 |
MAML3 |
mastermind-like 3 (Drosophila) |
46087 |
0.15 |
| chr8_77316854_77317005 | 0.04 |
ENSG00000222231 |
. |
138962 |
0.05 |
| chr2_176709130_176709424 | 0.04 |
AC016751.3 |
|
52787 |
0.15 |
| chr8_27975772_27976025 | 0.04 |
ELP3 |
elongator acetyltransferase complex subunit 3 |
24934 |
0.19 |
| chr9_126770993_126771177 | 0.04 |
RP11-85O21.2 |
|
590 |
0.64 |
| chr3_115867869_115868020 | 0.04 |
LSAMP-AS1 |
LSAMP antisense RNA 1 |
210927 |
0.02 |
| chr6_17968055_17968324 | 0.04 |
KIF13A |
kinesin family member 13A |
19505 |
0.26 |
| chr7_93552513_93552820 | 0.04 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
1655 |
0.33 |
| chr11_102158571_102158722 | 0.04 |
ENSG00000212466 |
. |
25807 |
0.15 |
| chr21_39630264_39630415 | 0.04 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
1469 |
0.53 |
| chr3_188077217_188077368 | 0.04 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
119640 |
0.06 |
| chr17_7905804_7906020 | 0.04 |
GUCY2D |
guanylate cyclase 2D, membrane (retina-specific) |
0 |
0.96 |
| chrX_47835789_47835940 | 0.04 |
ZNF182 |
zinc finger protein 182 |
27490 |
0.15 |
| chr5_173837713_173837864 | 0.04 |
CTC-229L21.1 |
|
237684 |
0.02 |
| chr8_41907411_41908748 | 0.04 |
KAT6A |
K(lysine) acetyltransferase 6A |
1426 |
0.43 |
| chr3_170807489_170807640 | 0.04 |
ENSG00000207963 |
. |
16984 |
0.23 |
| chrX_1600809_1601304 | 0.04 |
ASMTL |
acetylserotonin O-methyltransferase-like |
28401 |
0.16 |
| chr2_228680848_228680999 | 0.04 |
CCL20 |
chemokine (C-C motif) ligand 20 |
2353 |
0.32 |
| chr5_65749883_65750034 | 0.04 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
142218 |
0.05 |
| chr10_82266475_82266626 | 0.04 |
RP11-137H2.4 |
|
29148 |
0.16 |
| chr4_38080543_38080814 | 0.04 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
33186 |
0.23 |
| chr17_53361829_53361980 | 0.04 |
RP11-515O17.2 |
|
10890 |
0.24 |
| chr7_51383350_51383578 | 0.04 |
COBL |
cordon-bleu WH2 repeat protein |
704 |
0.82 |
| chr18_3794614_3794765 | 0.04 |
ENSG00000238790 |
. |
16628 |
0.17 |
| chr1_101468469_101468620 | 0.04 |
RP11-421L21.2 |
|
7515 |
0.15 |
| chr1_160676943_160677601 | 0.04 |
CD48 |
CD48 molecule |
4321 |
0.18 |
| chr7_373305_373494 | 0.04 |
AC187652.1 |
Protein LOC100996433 |
70481 |
0.11 |
| chr4_164614869_164615020 | 0.04 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
80258 |
0.11 |
| chr7_106358902_106359239 | 0.04 |
CTB-111H14.1 |
|
13825 |
0.24 |
| chr7_139637174_139637325 | 0.03 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
89869 |
0.08 |
| chr17_38255475_38255626 | 0.03 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
1428 |
0.29 |
| chr4_60684606_60684872 | 0.03 |
ENSG00000201676 |
. |
14154 |
0.24 |
| chr8_101259839_101259990 | 0.03 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
55544 |
0.1 |
| chr1_166895363_166895514 | 0.03 |
ILDR2 |
immunoglobulin-like domain containing receptor 2 |
49123 |
0.11 |
| chr4_169279306_169279573 | 0.03 |
DDX60 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
39481 |
0.15 |
| chr9_80596467_80596926 | 0.03 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
48824 |
0.19 |
| chr9_103192985_103193136 | 0.03 |
MSANTD3 |
Myb/SANT-like DNA-binding domain containing 3 |
3341 |
0.21 |
| chr6_133034700_133034851 | 0.03 |
VNN1 |
vanin 1 |
413 |
0.8 |
| chr2_100591103_100591254 | 0.03 |
AFF3 |
AF4/FMR2 family, member 3 |
129814 |
0.05 |
| chr1_21291714_21291865 | 0.03 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
7740 |
0.23 |
| chr8_133680386_133680537 | 0.03 |
LRRC6 |
leucine rich repeat containing 6 |
7344 |
0.21 |
| chr7_151039214_151039888 | 0.03 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
653 |
0.66 |
| chr7_26141990_26142141 | 0.03 |
ENSG00000266430 |
. |
42106 |
0.14 |
| chr6_49293859_49294324 | 0.03 |
ENSG00000252457 |
. |
18430 |
0.26 |
| chr7_20191764_20191915 | 0.03 |
MACC1-AS1 |
MACC1 antisense RNA 1 |
10300 |
0.2 |
| chr1_200877206_200877357 | 0.03 |
C1orf106 |
chromosome 1 open reading frame 106 |
13332 |
0.17 |
| chr7_143073130_143073319 | 0.03 |
AC093673.5 |
|
4364 |
0.12 |
| chr2_197180282_197180433 | 0.03 |
ENSG00000264627 |
. |
26224 |
0.18 |
| chr18_32621726_32622553 | 0.03 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
525 |
0.87 |
| chr22_23557480_23557631 | 0.03 |
BCR |
breakpoint cluster region |
35003 |
0.12 |
| chr20_62734157_62734308 | 0.03 |
NPBWR2 |
neuropeptides B/W receptor 2 |
4292 |
0.12 |
| chr12_10821249_10821400 | 0.03 |
STYK1 |
serine/threonine/tyrosine kinase 1 |
4933 |
0.2 |
| chr12_3226358_3226555 | 0.03 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
32507 |
0.17 |
| chr17_76210613_76211139 | 0.03 |
BIRC5 |
baculoviral IAP repeat containing 5 |
410 |
0.76 |
| chr11_63626215_63626366 | 0.03 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
8855 |
0.14 |
| chr17_46180907_46181094 | 0.03 |
SNX11 |
sorting nexin 11 |
281 |
0.82 |
| chr3_129512594_129512745 | 0.03 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
590 |
0.74 |
| chr3_32282148_32282299 | 0.03 |
CMTM8 |
CKLF-like MARVEL transmembrane domain containing 8 |
2052 |
0.26 |
| chr1_12495779_12495930 | 0.03 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
25930 |
0.2 |
| chr2_226896266_226896823 | 0.03 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
622947 |
0.0 |
| chrX_46510760_46510911 | 0.03 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
77616 |
0.09 |
| chr14_78073975_78074126 | 0.03 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
9066 |
0.2 |
| chr1_192128557_192128890 | 0.03 |
RGS18 |
regulator of G-protein signaling 18 |
1136 |
0.67 |
| chr9_41432727_41433091 | 0.03 |
SPATA31A5 |
SPATA31 subfamily A, member 5 |
67770 |
0.13 |
| chr11_100558368_100558828 | 0.03 |
CTD-2383M3.1 |
|
88 |
0.65 |
| chr13_80059310_80059736 | 0.03 |
NDFIP2 |
Nedd4 family interacting protein 2 |
3935 |
0.24 |
| chr18_77551163_77551314 | 0.03 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
72430 |
0.1 |
| chr21_33171558_33171709 | 0.03 |
SCAF4 |
SR-related CTD-associated factor 4 |
67245 |
0.11 |
| chr13_26623410_26623899 | 0.03 |
SHISA2 |
shisa family member 2 |
1515 |
0.52 |
| chr6_117804579_117805184 | 0.03 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
1056 |
0.49 |
| chr9_75767359_75768171 | 0.03 |
ANXA1 |
annexin A1 |
947 |
0.7 |
| chr1_28834546_28835031 | 0.03 |
ENSG00000200087 |
. |
283 |
0.74 |
| chr4_130264081_130264254 | 0.03 |
C4orf33 |
chromosome 4 open reading frame 33 |
246842 |
0.02 |
| chr10_5439032_5439183 | 0.03 |
TUBAL3 |
tubulin, alpha-like 3 |
7684 |
0.17 |
| chr18_60387000_60387290 | 0.03 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
4462 |
0.24 |
| chr7_28726429_28726958 | 0.03 |
CREB5 |
cAMP responsive element binding protein 5 |
1095 |
0.68 |
| chr12_15840984_15841159 | 0.03 |
ENSG00000200105 |
. |
16327 |
0.22 |
| chr3_127540841_127541975 | 0.03 |
MGLL |
monoglyceride lipase |
214 |
0.96 |
| chr2_216484448_216484719 | 0.03 |
AC012462.1 |
|
183607 |
0.03 |
| chr2_32233865_32234972 | 0.03 |
MEMO1 |
mediator of cell motility 1 |
1026 |
0.48 |
| chr2_208119475_208119962 | 0.03 |
AC007879.5 |
|
742 |
0.71 |
| chr3_63946362_63946540 | 0.03 |
ATXN7 |
ataxin 7 |
6969 |
0.17 |
| chr4_15900532_15900683 | 0.03 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
39364 |
0.15 |
| chr2_38055052_38055361 | 0.03 |
CDC42EP3 |
CDC42 effector protein (Rho GTPase binding) 3 |
89595 |
0.09 |
| chr3_177662383_177662611 | 0.03 |
ENSG00000199858 |
. |
63070 |
0.16 |
| chr3_69401946_69402170 | 0.03 |
FRMD4B |
FERM domain containing 4B |
790 |
0.78 |
| chr15_62464125_62464276 | 0.03 |
C2CD4B |
C2 calcium-dependent domain containing 4B |
6718 |
0.24 |
| chr3_98700853_98701271 | 0.03 |
ENSG00000207331 |
. |
73759 |
0.1 |
| chr3_150889747_150889898 | 0.03 |
ENSG00000199994 |
. |
16064 |
0.16 |
| chr4_183067512_183067663 | 0.03 |
AC108142.1 |
|
1185 |
0.34 |
| chr4_16034695_16034885 | 0.03 |
ENSG00000251758 |
. |
24380 |
0.19 |
| chr3_5303190_5303341 | 0.03 |
ENSG00000241227 |
. |
8357 |
0.2 |
| chr1_207226065_207226268 | 0.03 |
YOD1 |
YOD1 deubiquitinase |
159 |
0.77 |
| chr3_32464551_32464748 | 0.03 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
31118 |
0.19 |
| chr19_39184054_39184298 | 0.03 |
CTB-186G2.4 |
|
70 |
0.95 |
| chr3_57777559_57777710 | 0.03 |
SLMAP |
sarcolemma associated protein |
34349 |
0.18 |
| chr4_129523148_129523299 | 0.03 |
ENSG00000238802 |
. |
153958 |
0.04 |
| chr7_15279440_15279591 | 0.03 |
AGMO |
alkylglycerol monooxygenase |
126297 |
0.06 |
| chr1_103570260_103570411 | 0.03 |
COL11A1 |
collagen, type XI, alpha 1 |
3399 |
0.4 |
| chr17_75128680_75128831 | 0.03 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
4800 |
0.22 |
| chr17_79878072_79878223 | 0.03 |
SIRT7 |
sirtuin 7 |
1052 |
0.2 |
| chrY_1551045_1551310 | 0.03 |
NA |
NA |
> 106 |
NA |
| chr15_94841194_94841722 | 0.03 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
28 |
0.99 |
| chr9_5631788_5632177 | 0.03 |
KIAA1432 |
KIAA1432 |
2655 |
0.36 |
| chr16_19128205_19129316 | 0.03 |
ITPRIPL2 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
3506 |
0.19 |
| chrX_64921481_64921632 | 0.03 |
MSN |
moesin |
34019 |
0.23 |
| chr4_40567187_40567338 | 0.03 |
RBM47 |
RNA binding motif protein 47 |
49272 |
0.14 |
| chr16_85038693_85039085 | 0.03 |
ZDHHC7 |
zinc finger, DHHC-type containing 7 |
273 |
0.93 |
| chr9_39287322_39287607 | 0.03 |
CNTNAP3 |
contactin associated protein-like 3 |
628 |
0.81 |
| chr9_90118641_90118792 | 0.03 |
DAPK1 |
death-associated protein kinase 1 |
4796 |
0.3 |
| chr1_62752198_62752349 | 0.03 |
ENSG00000201153 |
. |
11655 |
0.2 |
| chr2_159947344_159947744 | 0.03 |
ENSG00000202029 |
. |
63890 |
0.13 |
| chr8_123875552_123875703 | 0.03 |
ZHX2 |
zinc fingers and homeoboxes 2 |
3 |
0.98 |
| chr11_46640743_46640894 | 0.03 |
HARBI1 |
harbinger transposase derived 1 |
1359 |
0.31 |
| chr7_33089731_33090334 | 0.03 |
ENSG00000241420 |
. |
4330 |
0.18 |
| chr2_207986359_207986603 | 0.03 |
KLF7-IT1 |
KLF7 intronic transcript 1 (non-protein coding) |
287 |
0.91 |
| chr2_189561015_189561166 | 0.03 |
DIRC1 |
disrupted in renal carcinoma 1 |
93741 |
0.08 |
| chr13_24007498_24007984 | 0.03 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
100 |
0.98 |
| chr9_136324511_136325310 | 0.03 |
CACFD1 |
calcium channel flower domain containing 1 |
179 |
0.91 |
| chr7_41530221_41530372 | 0.03 |
INHBA-AS1 |
INHBA antisense RNA 1 |
203218 |
0.03 |
| chr11_44647535_44647686 | 0.03 |
CD82 |
CD82 molecule |
7790 |
0.2 |
| chr18_60385543_60385694 | 0.03 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
2935 |
0.28 |
| chr1_55183020_55183171 | 0.03 |
TTC4 |
tetratricopeptide repeat domain 4 |
1600 |
0.36 |
| chr12_120978965_120979116 | 0.03 |
RNF10 |
ring finger protein 10 |
5811 |
0.12 |
| chr1_210412971_210413268 | 0.03 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
5727 |
0.24 |
| chr5_76171542_76171693 | 0.03 |
S100Z |
S100 calcium binding protein Z |
25693 |
0.15 |
| chr21_40179512_40179663 | 0.03 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
1712 |
0.5 |
| chrX_77198564_77198715 | 0.03 |
PGAM4 |
phosphoglycerate mutase family member 4 |
26496 |
0.14 |
| chr11_72451789_72452038 | 0.03 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
11535 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |