Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
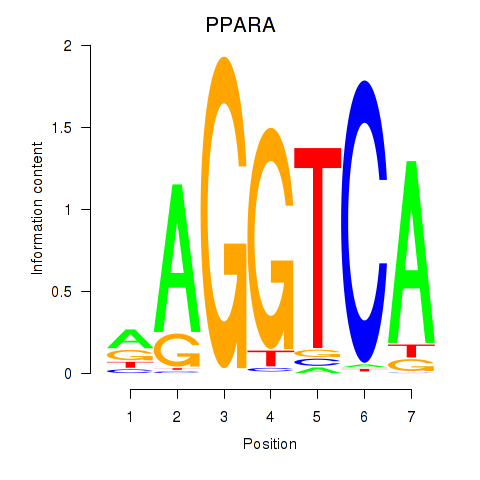
Results for PPARA
Z-value: 0.51
Transcription factors associated with PPARA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARA
|
ENSG00000186951.12 | peroxisome proliferator activated receptor alpha |
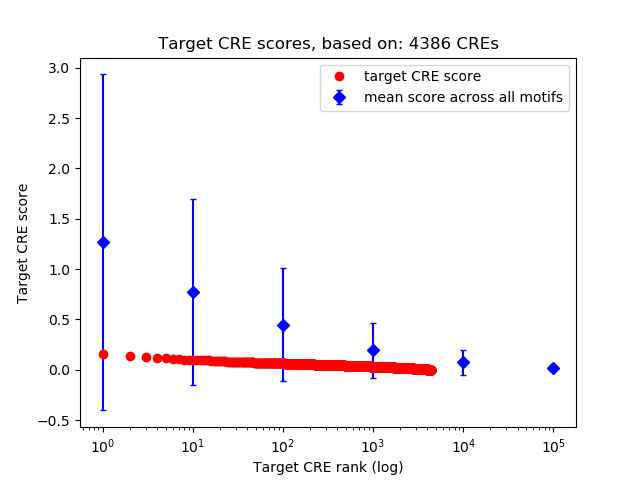
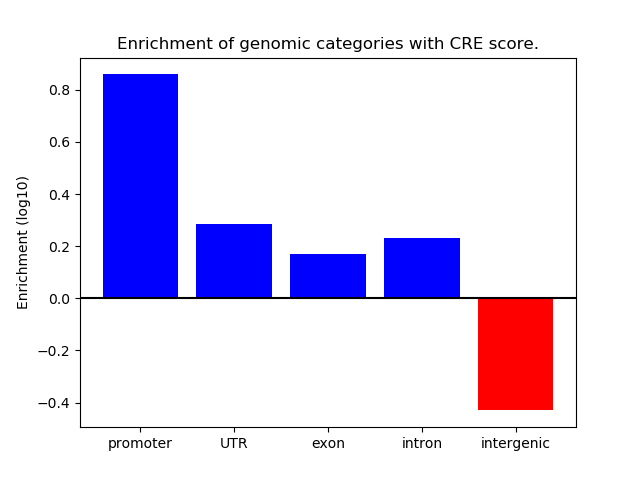
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr22_46544853_46545004 | PPARA | 1496 | 0.306121 | -0.32 | 4.0e-01 | Click! |
| chr22_46568570_46568721 | PPARA | 4000 | 0.183062 | -0.29 | 4.5e-01 | Click! |
| chr22_46544170_46544321 | PPARA | 2179 | 0.221745 | 0.27 | 4.8e-01 | Click! |
| chr22_46569183_46569334 | PPARA | 3387 | 0.196473 | 0.26 | 4.9e-01 | Click! |
| chr22_46546744_46547896 | PPARA | 467 | 0.747916 | 0.25 | 5.1e-01 | Click! |
Activity of the PPARA motif across conditions
Conditions sorted by the z-value of the PPARA motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
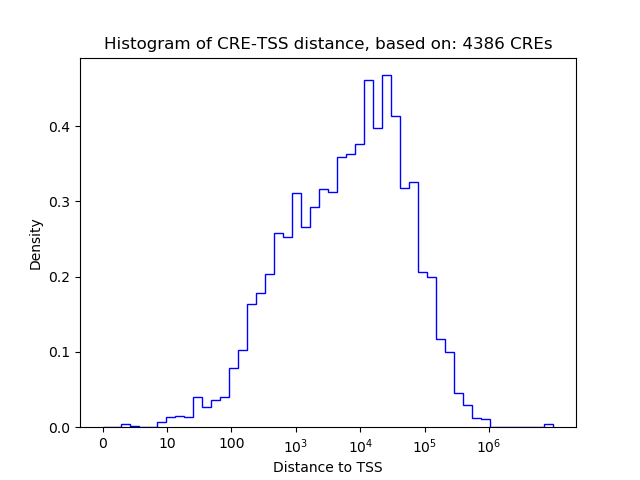
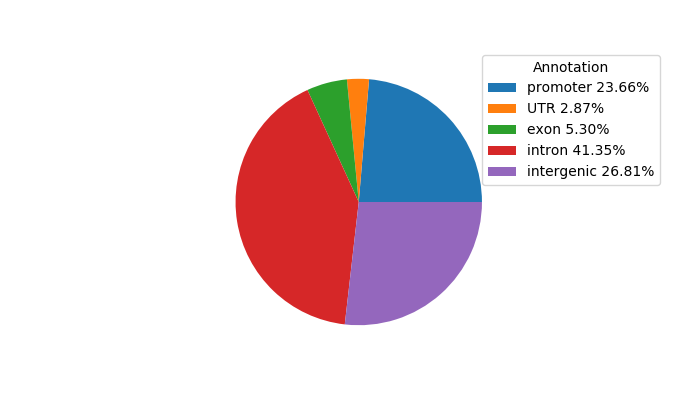
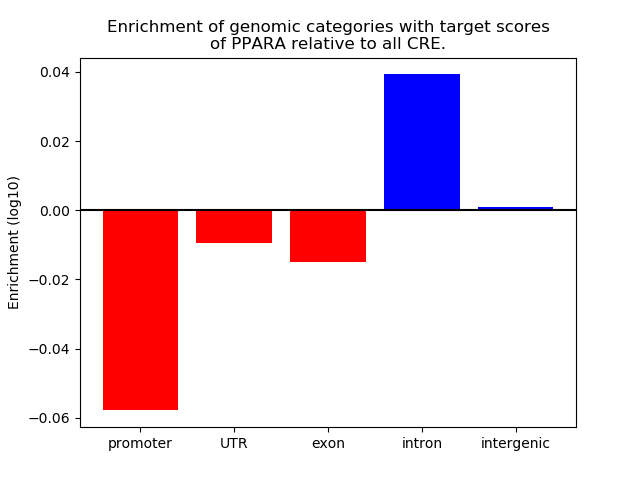
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_60273912_60274168 | 0.15 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
1140 |
0.65 |
| chr6_24743331_24743534 | 0.13 |
C6orf62 |
chromosome 6 open reading frame 62 |
22368 |
0.12 |
| chr16_89990293_89990742 | 0.13 |
TUBB3 |
Tubulin beta-3 chain |
743 |
0.44 |
| chr16_70718792_70718985 | 0.12 |
MTSS1L |
metastasis suppressor 1-like |
1081 |
0.43 |
| chr13_114864635_114864856 | 0.12 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
9550 |
0.23 |
| chr14_55117584_55117850 | 0.11 |
SAMD4A |
sterile alpha motif domain containing 4A |
83080 |
0.09 |
| chr11_789656_789830 | 0.11 |
CEND1 |
cell cycle exit and neuronal differentiation 1 |
380 |
0.63 |
| chr4_157805304_157805513 | 0.10 |
PDGFC |
platelet derived growth factor C |
33984 |
0.18 |
| chr6_11795100_11795449 | 0.10 |
ADTRP |
androgen-dependent TFPI-regulating protein |
12005 |
0.28 |
| chr5_169172915_169173066 | 0.10 |
DOCK2 |
dedicator of cytokinesis 2 |
13731 |
0.24 |
| chr15_60688043_60688242 | 0.10 |
ANXA2 |
annexin A2 |
1395 |
0.54 |
| chr19_44139197_44139620 | 0.10 |
CADM4 |
cell adhesion molecule 4 |
4583 |
0.1 |
| chr11_130319616_130319897 | 0.09 |
ADAMTS15 |
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
887 |
0.67 |
| chr12_56513564_56513771 | 0.09 |
RP11-603J24.6 |
|
56 |
0.9 |
| chr17_13265618_13265967 | 0.09 |
ENSG00000266115 |
. |
137853 |
0.05 |
| chr10_123871979_123872182 | 0.09 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
474 |
0.88 |
| chr17_41478521_41478741 | 0.09 |
ARL4D |
ADP-ribosylation factor-like 4D |
2304 |
0.18 |
| chr9_132429402_132429765 | 0.09 |
PRRX2 |
paired related homeobox 2 |
1663 |
0.28 |
| chr4_17782468_17782786 | 0.09 |
FAM184B |
family with sequence similarity 184, member B |
508 |
0.81 |
| chr4_184797459_184798247 | 0.09 |
STOX2 |
storkhead box 2 |
28656 |
0.22 |
| chr1_227503732_227504041 | 0.09 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
997 |
0.69 |
| chr5_90458282_90458577 | 0.09 |
ENSG00000199643 |
. |
108115 |
0.08 |
| chr3_159745160_159745451 | 0.09 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
11494 |
0.19 |
| chr20_43973524_43973810 | 0.08 |
SDC4 |
syndecan 4 |
3397 |
0.15 |
| chr15_48010788_48011252 | 0.08 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
334 |
0.93 |
| chr5_93222742_93222901 | 0.08 |
RP11-549J18.1 |
|
23290 |
0.26 |
| chr13_43843656_43843899 | 0.08 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
91436 |
0.1 |
| chr12_56236252_56236458 | 0.08 |
MMP19 |
matrix metallopeptidase 19 |
351 |
0.76 |
| chr3_12330541_12330692 | 0.08 |
PPARG |
peroxisome proliferator-activated receptor gamma |
46 |
0.99 |
| chr12_110720958_110721647 | 0.08 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
1816 |
0.41 |
| chr3_134091064_134091340 | 0.08 |
AMOTL2 |
angiomotin like 2 |
448 |
0.85 |
| chr12_118745072_118745584 | 0.08 |
TAOK3 |
TAO kinase 3 |
51464 |
0.15 |
| chr17_19967741_19968028 | 0.08 |
SPECC1 |
sperm antigen with calponin homology and coiled-coil domains 1 |
22450 |
0.18 |
| chr1_156092284_156092708 | 0.08 |
LMNA |
lamin A/C |
3455 |
0.14 |
| chr4_41215270_41215769 | 0.08 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
956 |
0.59 |
| chr1_119522601_119522807 | 0.08 |
TBX15 |
T-box 15 |
7724 |
0.28 |
| chrX_74967065_74967404 | 0.08 |
MAGEE2 |
melanoma antigen family E, 2 |
37845 |
0.23 |
| chr6_147236233_147236440 | 0.08 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
685 |
0.82 |
| chr13_52163965_52164370 | 0.08 |
ENSG00000206920 |
. |
3499 |
0.16 |
| chr15_30112674_30113382 | 0.08 |
TJP1 |
tight junction protein 1 |
723 |
0.64 |
| chr10_30238566_30238767 | 0.08 |
KIAA1462 |
KIAA1462 |
109787 |
0.07 |
| chr5_98397075_98397538 | 0.07 |
ENSG00000200351 |
. |
124855 |
0.06 |
| chr16_85205463_85205676 | 0.07 |
CTC-786C10.1 |
|
687 |
0.76 |
| chr9_113341179_113342041 | 0.07 |
SVEP1 |
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
213 |
0.96 |
| chr3_48646099_48647042 | 0.07 |
UQCRC1 |
ubiquinol-cytochrome c reductase core protein I |
900 |
0.38 |
| chr11_62162542_62162710 | 0.07 |
CTD-2531D15.5 |
|
1782 |
0.23 |
| chr4_146018335_146018486 | 0.07 |
ANAPC10 |
anaphase promoting complex subunit 10 |
124 |
0.9 |
| chr2_216298275_216298426 | 0.07 |
FN1 |
fibronectin 1 |
2440 |
0.3 |
| chr9_104248314_104249564 | 0.07 |
TMEM246 |
transmembrane protein 246 |
460 |
0.79 |
| chr6_81974819_81975050 | 0.07 |
RP1-300G12.2 |
|
269696 |
0.02 |
| chr2_227590884_227591406 | 0.07 |
ENSG00000263363 |
. |
67636 |
0.11 |
| chr10_105610252_105610446 | 0.07 |
SH3PXD2A |
SH3 and PX domains 2A |
4815 |
0.22 |
| chr1_163040363_163040678 | 0.07 |
RGS4 |
regulator of G-protein signaling 4 |
1314 |
0.56 |
| chr8_99956690_99957063 | 0.07 |
OSR2 |
odd-skipped related transciption factor 2 |
11 |
0.97 |
| chr22_46471692_46472277 | 0.07 |
FLJ27365 |
hsa-mir-4763 |
4208 |
0.11 |
| chr17_13503336_13503489 | 0.07 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
1832 |
0.45 |
| chr3_50194015_50194506 | 0.07 |
RP11-493K19.3 |
|
742 |
0.47 |
| chr15_35014576_35014814 | 0.07 |
GJD2 |
gap junction protein, delta 2, 36kDa |
32471 |
0.16 |
| chr7_95545859_95546042 | 0.07 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
112354 |
0.07 |
| chr1_120191467_120191618 | 0.07 |
ZNF697 |
zinc finger protein 697 |
1146 |
0.52 |
| chr19_3384704_3384855 | 0.07 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
18195 |
0.16 |
| chr2_190075356_190075623 | 0.07 |
COL5A2 |
collagen, type V, alpha 2 |
30884 |
0.2 |
| chr4_17809379_17809707 | 0.07 |
DCAF16 |
DDB1 and CUL4 associated factor 16 |
2838 |
0.25 |
| chr17_15181945_15182253 | 0.07 |
PMP22 |
peripheral myelin protein 22 |
13456 |
0.14 |
| chr1_27357912_27358708 | 0.07 |
FAM46B |
family with sequence similarity 46, member B |
18983 |
0.16 |
| chr7_73443076_73443679 | 0.07 |
ELN |
elastin |
859 |
0.62 |
| chr2_208378036_208378402 | 0.07 |
CREB1 |
cAMP responsive element binding protein 1 |
16242 |
0.2 |
| chr8_74333530_74333818 | 0.07 |
RP11-434I12.2 |
|
64978 |
0.13 |
| chrX_3204067_3204297 | 0.07 |
CXorf28 |
chromosome X open reading frame 28 |
14321 |
0.25 |
| chr18_52968140_52968291 | 0.07 |
TCF4 |
transcription factor 4 |
1642 |
0.52 |
| chr1_240760678_240760859 | 0.07 |
RP11-467I20.6 |
|
6720 |
0.21 |
| chr1_20125793_20126331 | 0.07 |
TMCO4 |
transmembrane and coiled-coil domains 4 |
225 |
0.92 |
| chr17_79315981_79316442 | 0.07 |
TMEM105 |
transmembrane protein 105 |
11737 |
0.13 |
| chr4_81198150_81198317 | 0.07 |
FGF5 |
fibroblast growth factor 5 |
10440 |
0.25 |
| chr6_58148368_58148782 | 0.07 |
ENSG00000212017 |
. |
893645 |
0.0 |
| chr7_15601189_15601685 | 0.07 |
AGMO |
alkylglycerol monooxygenase |
203 |
0.97 |
| chr17_1666629_1666885 | 0.07 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
634 |
0.58 |
| chr13_100752699_100753069 | 0.07 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
11547 |
0.27 |
| chr10_130991187_130991568 | 0.07 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
274071 |
0.02 |
| chr3_149374923_149375174 | 0.07 |
WWTR1 |
WW domain containing transcription regulator 1 |
174 |
0.54 |
| chr11_77733968_77734525 | 0.07 |
KCTD14 |
potassium channel tetramerization domain containing 14 |
94 |
0.72 |
| chr6_27205685_27206711 | 0.07 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chr15_68573463_68573639 | 0.07 |
FEM1B |
fem-1 homolog b (C. elegans) |
1254 |
0.47 |
| chr1_109247037_109247211 | 0.07 |
FNDC7 |
fibronectin type III domain containing 7 |
8155 |
0.16 |
| chr3_15677814_15677988 | 0.07 |
BTD |
biotinidase |
34425 |
0.13 |
| chr3_69888019_69888170 | 0.07 |
MITF |
microphthalmia-associated transcription factor |
27269 |
0.22 |
| chr3_16939119_16939271 | 0.07 |
PLCL2 |
phospholipase C-like 2 |
12743 |
0.25 |
| chr3_194592423_194592920 | 0.07 |
FAM43A |
family with sequence similarity 43, member A |
186049 |
0.02 |
| chr17_46561109_46561276 | 0.06 |
ENSG00000206805 |
. |
4733 |
0.14 |
| chr10_99441863_99442014 | 0.06 |
AVPI1 |
arginine vasopressin-induced 1 |
5142 |
0.16 |
| chr5_144624994_144625233 | 0.06 |
ENSG00000221467 |
. |
47775 |
0.2 |
| chr15_96883153_96883304 | 0.06 |
ENSG00000222651 |
. |
6738 |
0.16 |
| chr17_72868164_72868332 | 0.06 |
FDXR |
ferredoxin reductase |
838 |
0.46 |
| chr2_175863049_175863322 | 0.06 |
CHN1 |
chimerin 1 |
6769 |
0.16 |
| chr8_145022099_145022649 | 0.06 |
PLEC |
plectin |
1659 |
0.22 |
| chr4_166252557_166253372 | 0.06 |
MSMO1 |
methylsterol monooxygenase 1 |
3885 |
0.25 |
| chr20_10650822_10650973 | 0.06 |
RP11-103J8.1 |
|
2679 |
0.32 |
| chr3_8544328_8544516 | 0.06 |
LMCD1 |
LIM and cysteine-rich domains 1 |
850 |
0.49 |
| chr16_75150295_75150455 | 0.06 |
LDHD |
lactate dehydrogenase D |
290 |
0.87 |
| chr4_138531037_138531225 | 0.06 |
PCDH18 |
protocadherin 18 |
77483 |
0.13 |
| chr9_132949373_132949609 | 0.06 |
NCS1 |
neuronal calcium sensor 1 |
13381 |
0.21 |
| chrX_11456012_11456286 | 0.06 |
ARHGAP6 |
Rho GTPase activating protein 6 |
10256 |
0.3 |
| chr17_3379575_3379800 | 0.06 |
ASPA |
aspartoacylase |
391 |
0.8 |
| chr1_157980874_157981318 | 0.06 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
14244 |
0.2 |
| chr8_32406806_32407051 | 0.06 |
NRG1 |
neuregulin 1 |
683 |
0.82 |
| chr1_113931913_113932064 | 0.06 |
MAGI3 |
membrane associated guanylate kinase, WW and PDZ domain containing 3 |
1383 |
0.6 |
| chr15_74232964_74233156 | 0.06 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
12471 |
0.14 |
| chr15_52859194_52859345 | 0.06 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
1760 |
0.33 |
| chr19_48991098_48991290 | 0.06 |
CYTH2 |
cytohesin 2 |
15585 |
0.09 |
| chr10_93373658_93374073 | 0.06 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
18946 |
0.27 |
| chr12_110210896_110211119 | 0.06 |
FAM222A-AS1 |
FAM222A antisense RNA 1 |
282 |
0.9 |
| chr8_122648522_122648761 | 0.06 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
2892 |
0.31 |
| chr17_74697030_74697413 | 0.06 |
ENSG00000212418 |
. |
2588 |
0.13 |
| chr11_95888990_95889540 | 0.06 |
ENSG00000266192 |
. |
185337 |
0.03 |
| chr3_184037306_184037783 | 0.06 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
1 |
0.95 |
| chr20_54919132_54919450 | 0.06 |
FAM210B |
family with sequence similarity 210, member B |
14680 |
0.17 |
| chrX_115085279_115085526 | 0.06 |
ENSG00000266828 |
. |
23698 |
0.24 |
| chr3_167780902_167781122 | 0.06 |
GOLIM4 |
golgi integral membrane protein 4 |
32120 |
0.24 |
| chr13_52164714_52165080 | 0.06 |
ENSG00000242893 |
. |
3454 |
0.16 |
| chr16_30081137_30081369 | 0.06 |
ALDOA |
aldolase A, fructose-bisphosphate |
354 |
0.71 |
| chr1_41176173_41176466 | 0.06 |
NFYC |
nuclear transcription factor Y, gamma |
1104 |
0.44 |
| chr9_71746233_71746472 | 0.06 |
TJP2 |
tight junction protein 2 |
10128 |
0.26 |
| chr6_7468590_7468920 | 0.06 |
RP3-512B11.3 |
|
72816 |
0.09 |
| chr6_170053828_170054109 | 0.06 |
WDR27 |
WD repeat domain 27 |
6820 |
0.18 |
| chr6_17283021_17283300 | 0.06 |
RBM24 |
RNA binding motif protein 24 |
228 |
0.96 |
| chr6_26780154_26780414 | 0.06 |
GUSBP2 |
glucuronidase, beta pseudogene 2 |
66333 |
0.11 |
| chr3_143568633_143568784 | 0.06 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
1335 |
0.61 |
| chr7_101579659_101580126 | 0.06 |
CTB-181H17.1 |
|
23504 |
0.22 |
| chr11_707284_707470 | 0.06 |
EPS8L2 |
EPS8-like 2 |
748 |
0.42 |
| chr2_42599068_42599219 | 0.06 |
COX7A2L |
cytochrome c oxidase subunit VIIa polypeptide 2 like |
2993 |
0.32 |
| chr7_116141667_116142184 | 0.06 |
CAV2 |
caveolin 2 |
2091 |
0.24 |
| chr4_26789379_26789589 | 0.06 |
STIM2 |
stromal interaction molecule 2 |
69816 |
0.11 |
| chr5_17359988_17360246 | 0.06 |
ENSG00000201715 |
. |
14392 |
0.27 |
| chr3_116755540_116755902 | 0.06 |
ENSG00000200372 |
. |
71647 |
0.12 |
| chr10_30346435_30346778 | 0.06 |
KIAA1462 |
KIAA1462 |
1847 |
0.5 |
| chr14_86002689_86002854 | 0.06 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
6199 |
0.26 |
| chr6_106049688_106050083 | 0.06 |
PREP |
prolyl endopeptidase |
198926 |
0.03 |
| chr6_21808747_21809320 | 0.06 |
SOX4 |
SRY (sex determining region Y)-box 4 |
214962 |
0.02 |
| chr1_162601787_162601952 | 0.06 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
68 |
0.98 |
| chr6_158252834_158252985 | 0.06 |
RP11-52J3.3 |
|
1304 |
0.44 |
| chr11_118492062_118492969 | 0.06 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
14157 |
0.11 |
| chr7_17719632_17719796 | 0.06 |
SNX13 |
sorting nexin 13 |
260377 |
0.02 |
| chr6_28641788_28642196 | 0.06 |
ENSG00000272278 |
. |
25950 |
0.18 |
| chr15_74724718_74725429 | 0.06 |
SEMA7A |
semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) |
928 |
0.5 |
| chr6_26721817_26722019 | 0.06 |
ZNF322 |
zinc finger protein 322 |
61938 |
0.11 |
| chr21_35133624_35133954 | 0.06 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
26443 |
0.2 |
| chr8_19148668_19149118 | 0.06 |
SH2D4A |
SH2 domain containing 4A |
22235 |
0.28 |
| chr1_119524620_119524878 | 0.06 |
TBX15 |
T-box 15 |
5679 |
0.29 |
| chr2_181849023_181849317 | 0.06 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
2420 |
0.35 |
| chr5_102202832_102203172 | 0.06 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
1182 |
0.66 |
| chr6_27584810_27585278 | 0.06 |
ENSG00000238648 |
. |
14144 |
0.19 |
| chr19_17797231_17797557 | 0.06 |
UNC13A |
unc-13 homolog A (C. elegans) |
1614 |
0.31 |
| chr4_30722001_30722807 | 0.06 |
PCDH7 |
protocadherin 7 |
367 |
0.93 |
| chr6_150465056_150465588 | 0.06 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
1110 |
0.53 |
| chr1_12124202_12124372 | 0.06 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
760 |
0.56 |
| chr19_47366981_47367578 | 0.06 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
13030 |
0.17 |
| chr7_66532196_66532424 | 0.06 |
ENSG00000266525 |
. |
47074 |
0.12 |
| chr7_15723868_15724269 | 0.06 |
MEOX2 |
mesenchyme homeobox 2 |
2369 |
0.37 |
| chr12_96593279_96593430 | 0.06 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
4962 |
0.23 |
| chr17_76879212_76879961 | 0.06 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
9354 |
0.14 |
| chr12_52404839_52404990 | 0.06 |
GRASP |
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
53 |
0.96 |
| chr1_164724851_164725042 | 0.06 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
17203 |
0.18 |
| chr1_16030367_16030679 | 0.06 |
ENSG00000264048 |
. |
19556 |
0.09 |
| chr12_54417118_54417269 | 0.06 |
HOXC6 |
homeobox C6 |
4949 |
0.07 |
| chr6_27258471_27258730 | 0.06 |
POM121L2 |
POM121 transmembrane nucleoporin-like 2 |
20491 |
0.18 |
| chr4_20447726_20447877 | 0.06 |
SLIT2-IT1 |
SLIT2 intronic transcript 1 (non-protein coding) |
53989 |
0.15 |
| chr4_56731523_56731723 | 0.06 |
EXOC1 |
exocyst complex component 1 |
11718 |
0.19 |
| chr11_8832007_8832297 | 0.06 |
ST5 |
suppression of tumorigenicity 5 |
44 |
0.97 |
| chr6_116571521_116571672 | 0.06 |
TSPYL4 |
TSPY-like 4 |
3665 |
0.18 |
| chr1_161992740_161992891 | 0.06 |
OLFML2B |
olfactomedin-like 2B |
615 |
0.79 |
| chr1_8772875_8773203 | 0.06 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
9761 |
0.25 |
| chr5_11903634_11904112 | 0.06 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
237 |
0.97 |
| chr1_234792015_234792239 | 0.06 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
46856 |
0.14 |
| chr5_87436294_87437103 | 0.06 |
TMEM161B |
transmembrane protein 161B |
79750 |
0.11 |
| chr12_57611971_57612122 | 0.06 |
NXPH4 |
neurexophilin 4 |
1468 |
0.23 |
| chr12_15049535_15049773 | 0.06 |
MGP |
matrix Gla protein |
10794 |
0.15 |
| chr2_67601751_67601914 | 0.06 |
ETAA1 |
Ewing tumor-associated antigen 1 |
22619 |
0.29 |
| chr11_6341290_6341712 | 0.06 |
PRKCDBP |
protein kinase C, delta binding protein |
261 |
0.9 |
| chr8_132869766_132869917 | 0.06 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
46494 |
0.2 |
| chr5_53765392_53765543 | 0.06 |
HSPB3 |
heat shock 27kDa protein 3 |
14022 |
0.25 |
| chr2_3699150_3699650 | 0.06 |
ALLC |
allantoicase |
6385 |
0.16 |
| chr17_2869341_2869540 | 0.06 |
CTD-3060P21.1 |
|
251 |
0.94 |
| chr8_71580499_71580692 | 0.06 |
LACTB2 |
lactamase, beta 2 |
797 |
0.44 |
| chr19_11203405_11203676 | 0.06 |
LDLR |
low density lipoprotein receptor |
2265 |
0.24 |
| chr12_58819523_58819907 | 0.06 |
RP11-362K2.2 |
Protein LOC100506869 |
118192 |
0.06 |
| chr5_158520357_158520666 | 0.05 |
EBF1 |
early B-cell factor 1 |
6190 |
0.27 |
| chr8_32407781_32407984 | 0.05 |
NRG1 |
neuregulin 1 |
1637 |
0.54 |
| chr16_89392054_89392556 | 0.05 |
AC137932.6 |
|
2673 |
0.19 |
| chr15_96879714_96879865 | 0.05 |
ENSG00000222651 |
. |
3299 |
0.19 |
| chr6_27462859_27463076 | 0.05 |
ZNF184 |
zinc finger protein 184 |
22070 |
0.21 |
| chr8_27492286_27492503 | 0.05 |
SCARA3 |
scavenger receptor class A, member 3 |
696 |
0.66 |
| chrX_102940107_102940387 | 0.05 |
MORF4L2 |
mortality factor 4 like 2 |
902 |
0.43 |
| chr7_151433344_151433930 | 0.05 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
237 |
0.95 |
| chr10_4813272_4813482 | 0.05 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
15444 |
0.27 |
| chr1_161168438_161168589 | 0.05 |
ADAMTS4 |
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
330 |
0.57 |
| chr17_6451663_6451961 | 0.05 |
PITPNM3 |
PITPNM family member 3 |
7968 |
0.17 |
| chr18_77746974_77747125 | 0.05 |
TXNL4A |
thioredoxin-like 4A |
1488 |
0.39 |
| chr10_33248964_33249203 | 0.05 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
1923 |
0.46 |
| chr22_38145954_38146139 | 0.05 |
TRIOBP |
TRIO and F-actin binding protein |
958 |
0.43 |
| chr7_101525780_101526170 | 0.05 |
CTA-339C12.1 |
|
57966 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |