Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
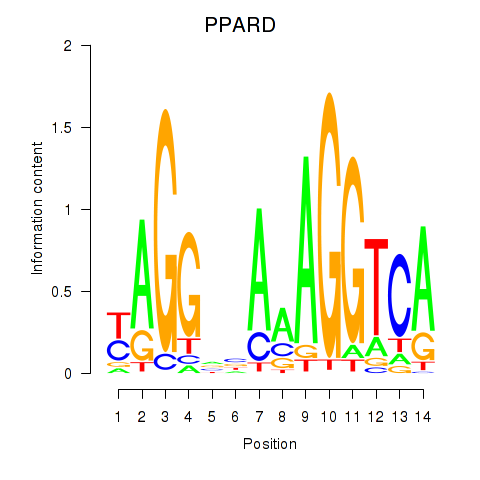
Results for PPARD
Z-value: 0.56
Transcription factors associated with PPARD
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PPARD
|
ENSG00000112033.9 | peroxisome proliferator activated receptor delta |
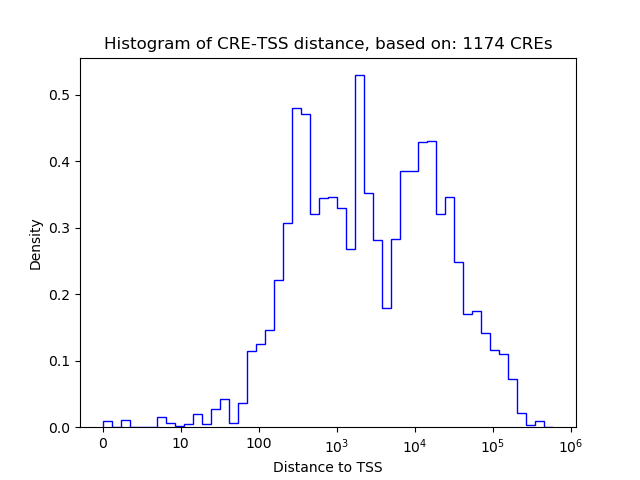
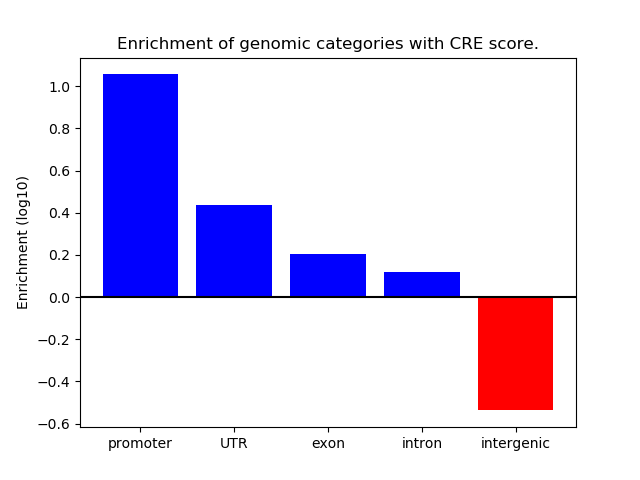
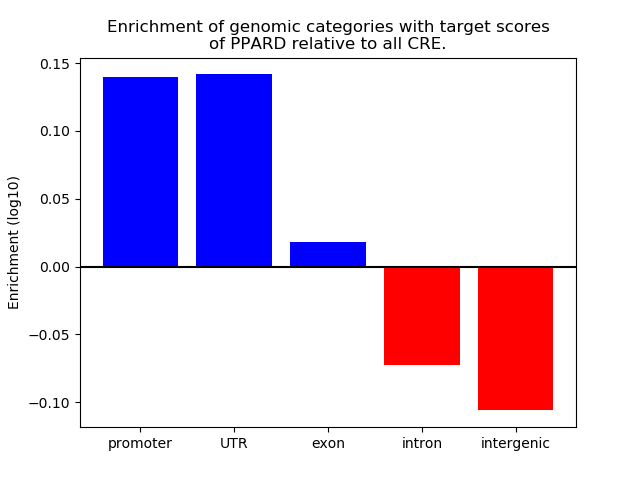
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_35310368_35310627 | PPARD | 95 | 0.970980 | 0.56 | 1.1e-01 | Click! |
| chr6_35353639_35354013 | PPARD | 43424 | 0.124590 | -0.48 | 1.9e-01 | Click! |
| chr6_35361239_35361390 | PPARD | 50912 | 0.107272 | -0.44 | 2.4e-01 | Click! |
| chr6_35348762_35348913 | PPARD | 38435 | 0.136611 | -0.44 | 2.4e-01 | Click! |
| chr6_35359348_35359522 | PPARD | 49033 | 0.111421 | -0.43 | 2.5e-01 | Click! |
Activity of the PPARD motif across conditions
Conditions sorted by the z-value of the PPARD motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
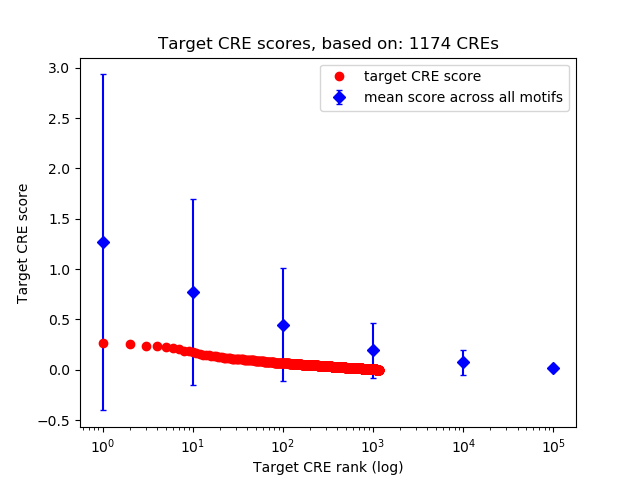
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_1990929_1991093 | 0.27 |
MSRB1 |
methionine sulfoxide reductase B1 |
2113 |
0.1 |
| chr7_139530944_139531095 | 0.25 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
1909 |
0.44 |
| chr8_33412041_33412395 | 0.24 |
RNF122 |
ring finger protein 122 |
12425 |
0.13 |
| chr7_127640981_127641152 | 0.23 |
LRRC4 |
leucine rich repeat containing 4 |
29992 |
0.21 |
| chr3_183273082_183273407 | 0.22 |
KLHL6 |
kelch-like family member 6 |
233 |
0.92 |
| chr2_182332192_182332343 | 0.22 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
10112 |
0.3 |
| chr9_88330487_88330638 | 0.21 |
AGTPBP1 |
ATP/GTP binding protein 1 |
22867 |
0.28 |
| chrX_70417643_70417794 | 0.19 |
RP5-1091N2.9 |
|
307 |
0.86 |
| chr15_77306736_77306887 | 0.19 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
1341 |
0.45 |
| chrX_117858717_117858999 | 0.18 |
IL13RA1 |
interleukin 13 receptor, alpha 1 |
2677 |
0.32 |
| chr7_6114490_6114641 | 0.16 |
ENSG00000264605 |
. |
8171 |
0.11 |
| chr14_73931523_73931791 | 0.16 |
ENSG00000251393 |
. |
2528 |
0.22 |
| chr1_160832331_160832502 | 0.15 |
CD244 |
CD244 molecule, natural killer cell receptor 2B4 |
74 |
0.97 |
| chr1_236056109_236056279 | 0.14 |
LYST |
lysosomal trafficking regulator |
9322 |
0.17 |
| chr15_49087139_49087290 | 0.14 |
RP11-485O10.2 |
|
11827 |
0.17 |
| chr1_26644198_26644349 | 0.14 |
CD52 |
CD52 molecule |
175 |
0.77 |
| chrX_47880068_47880219 | 0.14 |
SPACA5 |
sperm acrosome associated 5 |
12949 |
0.14 |
| chr12_10220623_10220774 | 0.14 |
ENSG00000223042 |
. |
16087 |
0.12 |
| chr2_28940107_28940258 | 0.13 |
AC097724.3 |
|
18793 |
0.15 |
| chr17_79879240_79879391 | 0.12 |
SIRT7 |
sirtuin 7 |
116 |
0.85 |
| chr11_67172347_67172531 | 0.12 |
TBC1D10C |
TBC1 domain family, member 10C |
779 |
0.35 |
| chr4_84033865_84034143 | 0.12 |
PLAC8 |
placenta-specific 8 |
1864 |
0.41 |
| chr15_57071498_57071649 | 0.12 |
ZNF280D |
zinc finger protein 280D |
45289 |
0.19 |
| chr16_70453211_70453362 | 0.11 |
ST3GAL2 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
18745 |
0.11 |
| chrX_118894160_118894311 | 0.11 |
SOWAHD |
sosondowah ankyrin repeat domain family member D |
1659 |
0.32 |
| chr6_33399521_33399672 | 0.11 |
SYNGAP1 |
synaptic Ras GTPase activating protein 1 |
88 |
0.94 |
| chr14_60621052_60621320 | 0.11 |
DHRS7 |
dehydrogenase/reductase (SDR family) member 7 |
1294 |
0.49 |
| chr15_83679938_83680110 | 0.11 |
C15orf40 |
chromosome 15 open reading frame 40 |
313 |
0.78 |
| chr3_72131377_72131562 | 0.11 |
ENSG00000212070 |
. |
180110 |
0.03 |
| chr7_105317322_105317473 | 0.11 |
ATXN7L1 |
ataxin 7-like 1 |
2212 |
0.41 |
| chr15_58438968_58439119 | 0.11 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
8019 |
0.23 |
| chr22_32033546_32033968 | 0.11 |
PISD |
phosphatidylserine decarboxylase |
634 |
0.74 |
| chr9_4760232_4760383 | 0.11 |
AK3 |
adenylate kinase 3 |
18264 |
0.17 |
| chr1_198898259_198898514 | 0.10 |
ENSG00000207759 |
. |
70104 |
0.12 |
| chr5_31361325_31361476 | 0.10 |
RP11-152K4.2 |
|
93683 |
0.08 |
| chr2_220112279_220112430 | 0.10 |
STK16 |
serine/threonine kinase 16 |
1736 |
0.14 |
| chr11_65656225_65656433 | 0.10 |
FIBP |
fibroblast growth factor (acidic) intracellular binding protein |
319 |
0.71 |
| chr15_45003998_45004149 | 0.10 |
B2M |
beta-2-microglobulin |
358 |
0.83 |
| chr2_216876893_216877044 | 0.10 |
MREG |
melanoregulin |
1378 |
0.5 |
| chr19_1068246_1068401 | 0.10 |
HMHA1 |
histocompatibility (minor) HA-1 |
826 |
0.4 |
| chrX_153659493_153659733 | 0.10 |
ATP6AP1 |
ATPase, H+ transporting, lysosomal accessory protein 1 |
2604 |
0.11 |
| chr20_54989282_54989551 | 0.10 |
CASS4 |
Cas scaffolding protein family member 4 |
2099 |
0.24 |
| chr11_74974835_74974986 | 0.10 |
CTD-2562J17.9 |
|
423 |
0.77 |
| chr2_26679414_26679565 | 0.10 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
8322 |
0.2 |
| chr2_127421217_127421368 | 0.10 |
GYPC |
glycophorin C (Gerbich blood group) |
7532 |
0.28 |
| chr9_4740257_4740758 | 0.10 |
AK3 |
adenylate kinase 3 |
720 |
0.68 |
| chrX_77359847_77360362 | 0.10 |
PGK1 |
phosphoglycerate kinase 1 |
352 |
0.91 |
| chr2_113596772_113596923 | 0.09 |
IL1B |
interleukin 1, beta |
2367 |
0.26 |
| chr11_61733702_61734635 | 0.09 |
FTH1 |
ferritin, heavy polypeptide 1 |
470 |
0.67 |
| chr15_70379620_70379771 | 0.09 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
7429 |
0.24 |
| chr8_103666291_103666442 | 0.09 |
KLF10 |
Kruppel-like factor 10 |
237 |
0.95 |
| chr10_30831582_30831733 | 0.09 |
ENSG00000239744 |
. |
13176 |
0.24 |
| chrX_117632028_117632179 | 0.09 |
DOCK11 |
dedicator of cytokinesis 11 |
2231 |
0.39 |
| chr7_105923338_105923489 | 0.09 |
NAMPT |
nicotinamide phosphoribosyltransferase |
1954 |
0.34 |
| chr17_30816403_30816586 | 0.09 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
1680 |
0.24 |
| chrX_71269220_71269371 | 0.09 |
RGAG4 |
retrotransposon gag domain containing 4 |
82383 |
0.07 |
| chr16_81944390_81944667 | 0.09 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
27654 |
0.19 |
| chr8_29356447_29357119 | 0.09 |
RP4-676L2.1 |
|
146096 |
0.04 |
| chr5_169705024_169705175 | 0.08 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
10768 |
0.22 |
| chr6_49603951_49604102 | 0.08 |
RHAG |
Rh-associated glycoprotein |
526 |
0.83 |
| chr19_58315098_58315340 | 0.08 |
ZNF552 |
zinc finger protein 552 |
9553 |
0.1 |
| chr2_97466623_97466774 | 0.08 |
ENSG00000264157 |
. |
2683 |
0.2 |
| chrX_118603274_118603731 | 0.08 |
SLC25A5 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 |
1139 |
0.39 |
| chr4_100815773_100816143 | 0.08 |
LAMTOR3 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
311 |
0.91 |
| chr1_161038546_161039545 | 0.08 |
ARHGAP30 |
Rho GTPase activating protein 30 |
411 |
0.65 |
| chr11_67171589_67172183 | 0.08 |
TBC1D10C |
TBC1 domain family, member 10C |
226 |
0.8 |
| chr20_35273778_35274551 | 0.08 |
SLA2 |
Src-like-adaptor 2 |
120 |
0.95 |
| chr9_80524074_80524374 | 0.08 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86309 |
0.1 |
| chrX_45050678_45050829 | 0.08 |
RP11-342D14.1 |
|
8257 |
0.24 |
| chr20_4807814_4807965 | 0.08 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
3598 |
0.24 |
| chr1_36616449_36616600 | 0.08 |
TRAPPC3 |
trafficking protein particle complex 3 |
1426 |
0.33 |
| chr9_70850762_70850954 | 0.08 |
CBWD3 |
COBW domain containing 3 |
5539 |
0.19 |
| chr11_1781045_1781196 | 0.08 |
AC068580.6 |
|
458 |
0.52 |
| chr11_3886844_3887200 | 0.08 |
STIM1 |
stromal interaction molecule 1 |
1311 |
0.29 |
| chr7_134832355_134832754 | 0.08 |
TMEM140 |
transmembrane protein 140 |
270 |
0.58 |
| chr7_642417_642568 | 0.08 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
172 |
0.69 |
| chr9_90117020_90117171 | 0.07 |
DAPK1 |
death-associated protein kinase 1 |
3175 |
0.34 |
| chrX_48772131_48772362 | 0.07 |
PIM2 |
pim-2 oncogene |
766 |
0.45 |
| chr14_52513608_52513759 | 0.07 |
NID2 |
nidogen 2 (osteonidogen) |
8013 |
0.21 |
| chr17_18873108_18873266 | 0.07 |
FAM83G |
family with sequence similarity 83, member G |
8014 |
0.12 |
| chr2_175532806_175532957 | 0.07 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
14718 |
0.23 |
| chr9_69268507_69268733 | 0.07 |
CBWD6 |
COBW domain containing 6 |
6027 |
0.21 |
| chr16_50751041_50751192 | 0.07 |
RP11-327F22.6 |
|
4361 |
0.13 |
| chr19_43849967_43850118 | 0.07 |
CD177 |
CD177 molecule |
7783 |
0.17 |
| chr2_48133928_48134737 | 0.07 |
AC079807.2 |
|
363 |
0.84 |
| chr16_22200588_22200739 | 0.07 |
EEF2K |
eukaryotic elongation factor-2 kinase |
16940 |
0.16 |
| chr1_33458604_33458774 | 0.07 |
RP1-117O3.2 |
|
6013 |
0.15 |
| chr7_95238082_95238332 | 0.07 |
AC002451.3 |
|
901 |
0.62 |
| chr4_159593559_159594079 | 0.07 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
328 |
0.69 |
| chr2_54816430_54816697 | 0.07 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
31032 |
0.16 |
| chr1_172870111_172870262 | 0.07 |
TNFSF18 |
tumor necrosis factor (ligand) superfamily, member 18 |
149870 |
0.04 |
| chr8_1953776_1953973 | 0.07 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
31830 |
0.2 |
| chr5_40384992_40385143 | 0.07 |
ENSG00000265615 |
. |
64647 |
0.14 |
| chr9_33165755_33166165 | 0.07 |
RP11-326F20.5 |
|
1013 |
0.39 |
| chr1_26799708_26800405 | 0.07 |
HMGN2 |
high mobility group nucleosomal binding domain 2 |
1046 |
0.42 |
| chr20_1926522_1926673 | 0.07 |
RP4-684O24.5 |
|
1295 |
0.5 |
| chr10_106083791_106084156 | 0.07 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
4709 |
0.15 |
| chr6_71998977_71999172 | 0.07 |
OGFRL1 |
opioid growth factor receptor-like 1 |
568 |
0.78 |
| chr9_139928371_139928559 | 0.07 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
1003 |
0.23 |
| chr11_112030056_112030207 | 0.07 |
IL18 |
interleukin 18 (interferon-gamma-inducing factor) |
4666 |
0.12 |
| chr2_620261_620412 | 0.07 |
TMEM18 |
transmembrane protein 18 |
55439 |
0.13 |
| chr3_196337100_196337251 | 0.07 |
LINC01063 |
long intergenic non-protein coding RNA 1063 |
22283 |
0.11 |
| chr14_24805236_24805387 | 0.07 |
ADCY4 |
adenylate cyclase 4 |
1034 |
0.21 |
| chr11_47415173_47415381 | 0.07 |
RP11-750H9.5 |
|
1914 |
0.17 |
| chr5_58958074_58958460 | 0.07 |
ENSG00000202601 |
. |
41262 |
0.2 |
| chr11_128585758_128586117 | 0.07 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
20019 |
0.17 |
| chr19_4069446_4069597 | 0.07 |
ZBTB7A |
zinc finger and BTB domain containing 7A |
2578 |
0.17 |
| chr9_102587867_102588202 | 0.06 |
NR4A3 |
nuclear receptor subfamily 4, group A, member 3 |
975 |
0.66 |
| chr2_136690107_136690258 | 0.06 |
DARS |
aspartyl-tRNA synthetase |
12059 |
0.21 |
| chrX_12995628_12996058 | 0.06 |
TMSB4X |
thymosin beta 4, X-linked |
2066 |
0.37 |
| chr12_95037835_95038747 | 0.06 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
6047 |
0.29 |
| chr10_73520322_73520473 | 0.06 |
C10orf54 |
chromosome 10 open reading frame 54 |
3010 |
0.26 |
| chr15_40393037_40393188 | 0.06 |
BMF |
Bcl2 modifying factor |
5175 |
0.17 |
| chr1_17630308_17630459 | 0.06 |
PADI4 |
peptidyl arginine deiminase, type IV |
4307 |
0.19 |
| chr13_32606969_32607251 | 0.06 |
FRY-AS1 |
FRY antisense RNA 1 |
1334 |
0.41 |
| chr19_45394510_45394714 | 0.06 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
118 |
0.81 |
| chr1_203924787_203924938 | 0.06 |
ENSG00000211554 |
. |
43609 |
0.15 |
| chr15_69606581_69606732 | 0.06 |
PAQR5 |
progestin and adipoQ receptor family member V |
86 |
0.96 |
| chr6_13274566_13274804 | 0.06 |
RP1-257A7.4 |
|
394 |
0.82 |
| chr8_68880458_68880609 | 0.06 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
16180 |
0.28 |
| chr19_2428027_2428178 | 0.06 |
TIMM13 |
translocase of inner mitochondrial membrane 13 homolog (yeast) |
210 |
0.91 |
| chr9_126138360_126138511 | 0.06 |
CRB2 |
crumbs homolog 2 (Drosophila) |
7267 |
0.23 |
| chr15_52843571_52843722 | 0.06 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
6295 |
0.19 |
| chr8_49296754_49296905 | 0.06 |
ENSG00000252710 |
. |
76239 |
0.12 |
| chr21_39869780_39870199 | 0.06 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
356 |
0.93 |
| chr1_184834953_184835104 | 0.06 |
ENSG00000252222 |
. |
44405 |
0.16 |
| chr6_43742717_43743167 | 0.06 |
VEGFA |
vascular endothelial growth factor A |
852 |
0.59 |
| chr9_92756542_92756693 | 0.06 |
ENSG00000263967 |
. |
29200 |
0.27 |
| chrX_70330282_70330478 | 0.06 |
IL2RG |
interleukin 2 receptor, gamma |
368 |
0.76 |
| chr11_65191721_65192304 | 0.06 |
ENSG00000245532 |
. |
19917 |
0.1 |
| chr20_1797966_1798117 | 0.06 |
SIRPA |
signal-regulatory protein alpha |
77113 |
0.09 |
| chr19_43891664_43891815 | 0.06 |
TEX101 |
testis expressed 101 |
13909 |
0.15 |
| chr5_175960940_175961324 | 0.06 |
RNF44 |
ring finger protein 44 |
3289 |
0.16 |
| chr1_198645627_198645927 | 0.06 |
RP11-553K8.5 |
|
9587 |
0.26 |
| chr10_103986046_103986414 | 0.06 |
ELOVL3 |
ELOVL fatty acid elongase 3 |
145 |
0.94 |
| chr1_231750656_231750807 | 0.06 |
LINC00582 |
long intergenic non-protein coding RNA 582 |
2895 |
0.26 |
| chr7_1067729_1067964 | 0.06 |
C7orf50 |
chromosome 7 open reading frame 50 |
129 |
0.88 |
| chr6_2837966_2838179 | 0.06 |
SERPINB1 |
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
4022 |
0.22 |
| chr17_39464324_39464480 | 0.06 |
KRTAP16-1 |
keratin associated protein 16-1 |
1103 |
0.28 |
| chr1_11786795_11786946 | 0.06 |
AGTRAP |
angiotensin II receptor-associated protein |
9271 |
0.12 |
| chr3_36937722_36937873 | 0.06 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
37423 |
0.17 |
| chr13_51168043_51168194 | 0.06 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
213874 |
0.02 |
| chr18_2637852_2638003 | 0.06 |
ENSG00000200875 |
. |
11939 |
0.14 |
| chrX_118894330_118894481 | 0.06 |
SOWAHD |
sosondowah ankyrin repeat domain family member D |
1829 |
0.3 |
| chr7_127749300_127749451 | 0.06 |
ENSG00000207588 |
. |
27462 |
0.2 |
| chr16_67222162_67222313 | 0.06 |
EXOC3L1 |
exocyst complex component 3-like 1 |
1802 |
0.13 |
| chr13_25173335_25173622 | 0.05 |
ENSG00000211508 |
. |
9739 |
0.19 |
| chr1_169861896_169862644 | 0.05 |
SCYL3 |
SCY1-like 3 (S. cerevisiae) |
806 |
0.65 |
| chr18_2984866_2985017 | 0.05 |
LPIN2 |
lipin 2 |
2070 |
0.27 |
| chr9_115142438_115143003 | 0.05 |
HSDL2 |
hydroxysteroid dehydrogenase like 2 |
328 |
0.91 |
| chr12_125412865_125413207 | 0.05 |
UBC |
ubiquitin C |
11122 |
0.16 |
| chr1_159054629_159054780 | 0.05 |
AIM2 |
absent in melanoma 2 |
8013 |
0.19 |
| chr15_93426430_93426631 | 0.05 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
4 |
0.98 |
| chr1_244997779_244998465 | 0.05 |
COX20 |
COX20 cytochrome C oxidase assembly factor |
502 |
0.79 |
| chr19_38806035_38806435 | 0.05 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
173 |
0.87 |
| chr7_38314416_38314567 | 0.05 |
STARD3NL |
STARD3 N-terminal like |
96494 |
0.09 |
| chr22_40795827_40795978 | 0.05 |
SGSM3 |
small G protein signaling modulator 3 |
8415 |
0.15 |
| chr20_52240537_52240725 | 0.05 |
ZNF217 |
zinc finger protein 217 |
15200 |
0.2 |
| chr6_109119373_109119524 | 0.05 |
ARMC2 |
armadillo repeat containing 2 |
50171 |
0.16 |
| chr4_6927779_6927930 | 0.05 |
TBC1D14 |
TBC1 domain family, member 14 |
15879 |
0.16 |
| chr8_56756788_56757159 | 0.05 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
35399 |
0.12 |
| chr17_80476965_80477437 | 0.05 |
FOXK2 |
forkhead box K2 |
390 |
0.76 |
| chr19_7268349_7268500 | 0.05 |
INSR |
insulin receptor |
25518 |
0.17 |
| chr20_34329591_34330097 | 0.05 |
RBM39 |
RNA binding motif protein 39 |
29 |
0.96 |
| chr10_129678816_129678967 | 0.05 |
CLRN3 |
clarin 3 |
12320 |
0.22 |
| chr6_111180428_111180655 | 0.05 |
ENSG00000199360 |
. |
2921 |
0.21 |
| chr9_108612973_108613124 | 0.05 |
TMEM38B |
transmembrane protein 38B |
129080 |
0.06 |
| chr10_89338330_89338481 | 0.05 |
ENSG00000222192 |
. |
15462 |
0.22 |
| chr7_157182579_157182895 | 0.05 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
50222 |
0.15 |
| chr12_53759890_53760041 | 0.05 |
SP1 |
Sp1 transcription factor |
13995 |
0.11 |
| chrX_38494089_38494240 | 0.05 |
ENSG00000238606 |
. |
26696 |
0.23 |
| chr1_101004250_101004401 | 0.05 |
GPR88 |
G protein-coupled receptor 88 |
632 |
0.79 |
| chr7_76033306_76033511 | 0.05 |
SRCRB4D |
scavenger receptor cysteine rich domain containing, group B (4 domains) |
5604 |
0.17 |
| chr9_139927070_139927409 | 0.05 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
223 |
0.78 |
| chr7_112089913_112090222 | 0.05 |
IFRD1 |
interferon-related developmental regulator 1 |
37 |
0.98 |
| chr17_7462411_7462570 | 0.05 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
386 |
0.58 |
| chr3_46973710_46973887 | 0.05 |
PTH1R |
parathyroid hormone 1 receptor |
29709 |
0.12 |
| chr19_18334931_18335362 | 0.05 |
PDE4C |
phosphodiesterase 4C, cAMP-specific |
157 |
0.91 |
| chr15_63486470_63486758 | 0.05 |
RAB8B |
RAB8B, member RAS oncogene family |
4809 |
0.23 |
| chr7_38313129_38313494 | 0.05 |
STARD3NL |
STARD3 N-terminal like |
95314 |
0.09 |
| chr7_72936777_72937003 | 0.05 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
282 |
0.88 |
| chr2_109237700_109238010 | 0.05 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
133 |
0.97 |
| chr3_44551397_44551548 | 0.05 |
ZNF852 |
zinc finger protein 852 |
656 |
0.63 |
| chr10_134364825_134364982 | 0.05 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
13260 |
0.21 |
| chr22_41253195_41254018 | 0.05 |
XPNPEP3 |
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
504 |
0.47 |
| chr8_61823770_61824122 | 0.05 |
RP11-33I11.2 |
|
101781 |
0.08 |
| chr2_197024297_197024448 | 0.05 |
STK17B |
serine/threonine kinase 17b |
3001 |
0.26 |
| chr1_206254058_206254209 | 0.05 |
ENSG00000252692 |
. |
6960 |
0.17 |
| chr13_99930202_99930387 | 0.05 |
GPR18 |
G protein-coupled receptor 18 |
16296 |
0.18 |
| chr11_67043892_67044043 | 0.05 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
10015 |
0.11 |
| chr20_35898453_35898604 | 0.05 |
GHRH |
growth hormone releasing hormone |
8290 |
0.19 |
| chr16_30393182_30393494 | 0.05 |
SEPT1 |
septin 1 |
414 |
0.64 |
| chr11_48055572_48055723 | 0.05 |
AC103828.1 |
|
18240 |
0.2 |
| chr17_2027532_2027720 | 0.05 |
RP11-667K14.5 |
|
34651 |
0.09 |
| chr14_93651605_93652048 | 0.05 |
TMEM251 |
transmembrane protein 251 |
468 |
0.34 |
| chr7_4681559_4681752 | 0.05 |
FOXK1 |
forkhead box K1 |
1733 |
0.44 |
| chr17_27466417_27467105 | 0.05 |
MYO18A |
myosin XVIIIA |
675 |
0.43 |
| chr11_126238269_126238471 | 0.05 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
12501 |
0.16 |
| chr12_2921492_2921671 | 0.05 |
RP4-816N1.6 |
|
34 |
0.71 |
| chr12_57030254_57030540 | 0.05 |
BAZ2A |
bromodomain adjacent to zinc finger domain, 2A |
234 |
0.88 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0006863 | purine nucleobase transport(GO:0006863) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |