Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
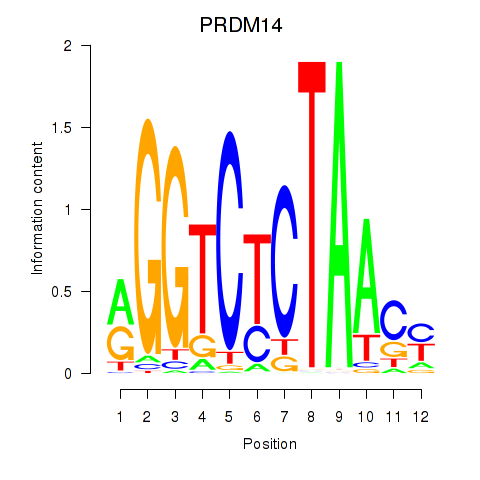
Results for PRDM14
Z-value: 0.90
Transcription factors associated with PRDM14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM14
|
ENSG00000147596.3 | PR/SET domain 14 |
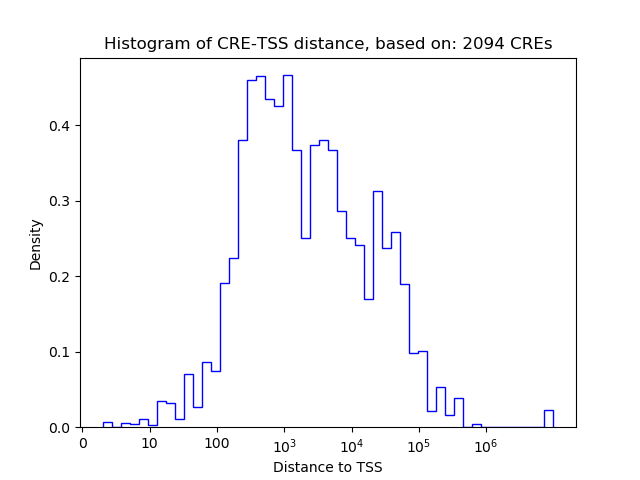
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_70984934_70985085 | PRDM14 | 1081 | 0.541383 | -0.60 | 8.5e-02 | Click! |
| chr8_70984555_70984706 | PRDM14 | 702 | 0.707363 | -0.55 | 1.2e-01 | Click! |
| chr8_70982226_70982377 | PRDM14 | 1261 | 0.490365 | 0.46 | 2.1e-01 | Click! |
| chr8_70947239_70947390 | PRDM14 | 36248 | 0.155587 | 0.13 | 7.4e-01 | Click! |
| chr8_70984004_70984275 | PRDM14 | 211 | 0.944506 | 0.03 | 9.5e-01 | Click! |
Activity of the PRDM14 motif across conditions
Conditions sorted by the z-value of the PRDM14 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
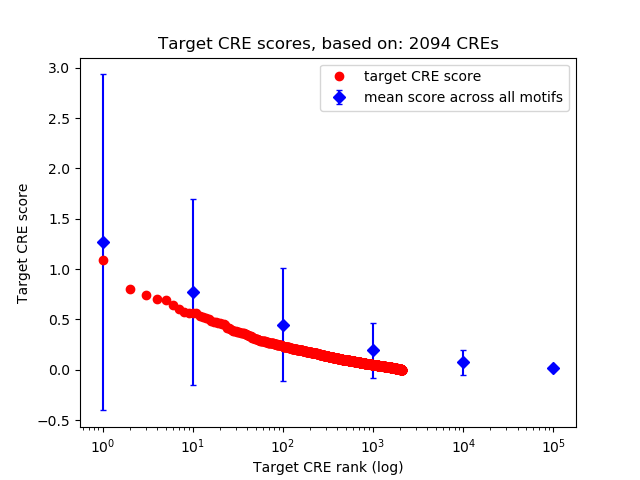
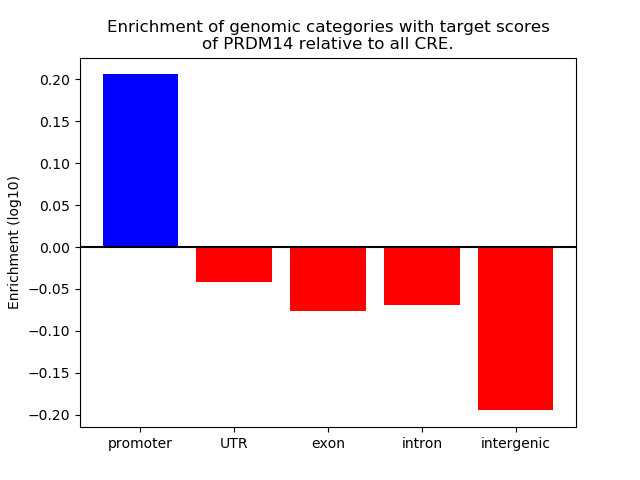
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr22_40297430_40297673 | 1.09 |
GRAP2 |
GRB2-related adaptor protein 2 |
438 |
0.81 |
| chr4_109085952_109086103 | 0.80 |
LEF1 |
lymphoid enhancer-binding factor 1 |
1430 |
0.44 |
| chr6_501893_502410 | 0.74 |
RP1-20B11.2 |
|
22020 |
0.26 |
| chr5_133455627_133455909 | 0.71 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
3541 |
0.27 |
| chr10_11208288_11208755 | 0.69 |
CELF2 |
CUGBP, Elav-like family member 2 |
1030 |
0.57 |
| chr1_118149801_118150025 | 0.64 |
FAM46C |
family with sequence similarity 46, member C |
1357 |
0.38 |
| chr14_22993528_22993679 | 0.60 |
TRAJ15 |
T cell receptor alpha joining 15 |
4977 |
0.12 |
| chr15_86126717_86127003 | 0.58 |
RP11-815J21.2 |
|
3451 |
0.22 |
| chr8_134509191_134509342 | 0.57 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
2338 |
0.44 |
| chr5_133454026_133454256 | 0.57 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
2825 |
0.3 |
| chr17_78845601_78845793 | 0.56 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
50827 |
0.1 |
| chr17_38716835_38717173 | 0.53 |
CCR7 |
chemokine (C-C motif) receptor 7 |
261 |
0.91 |
| chr20_56202934_56203085 | 0.53 |
ZBP1 |
Z-DNA binding protein 1 |
7377 |
0.24 |
| chr10_82228022_82228360 | 0.51 |
TSPAN14 |
tetraspanin 14 |
9133 |
0.21 |
| chr15_75080868_75081404 | 0.50 |
ENSG00000264386 |
. |
38 |
0.76 |
| chr2_27072177_27072340 | 0.49 |
DPYSL5 |
dihydropyrimidinase-like 5 |
882 |
0.61 |
| chr1_160737807_160738093 | 0.48 |
LY9 |
lymphocyte antigen 9 |
27914 |
0.13 |
| chr17_80274181_80274428 | 0.48 |
CD7 |
CD7 molecule |
1124 |
0.35 |
| chr19_1029334_1029485 | 0.47 |
CNN2 |
calponin 2 |
1727 |
0.16 |
| chr12_51318719_51318989 | 0.47 |
METTL7A |
methyltransferase like 7A |
320 |
0.86 |
| chr6_2222017_2222664 | 0.46 |
GMDS |
GDP-mannose 4,6-dehydratase |
23586 |
0.28 |
| chr9_6414096_6414470 | 0.45 |
UHRF2 |
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
865 |
0.64 |
| chr6_24910150_24910301 | 0.45 |
FAM65B |
family with sequence similarity 65, member B |
970 |
0.62 |
| chr20_822528_822694 | 0.42 |
FAM110A |
family with sequence similarity 110, member A |
2674 |
0.33 |
| chr8_30776682_30776833 | 0.42 |
TEX15 |
testis expressed 15 |
70149 |
0.11 |
| chr9_96341921_96342072 | 0.40 |
PHF2 |
PHD finger protein 2 |
3087 |
0.26 |
| chr11_46353937_46354088 | 0.40 |
DGKZ |
diacylglycerol kinase, zeta |
443 |
0.79 |
| chr3_46995163_46995374 | 0.39 |
CCDC12 |
coiled-coil domain containing 12 |
23002 |
0.14 |
| chr6_32605503_32605654 | 0.39 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
381 |
0.81 |
| chr2_182322868_182323440 | 0.38 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
999 |
0.69 |
| chr4_109062684_109062835 | 0.38 |
LEF1 |
lymphoid enhancer-binding factor 1 |
24698 |
0.2 |
| chr6_159465648_159465970 | 0.38 |
TAGAP |
T-cell activation RhoGTPase activating protein |
241 |
0.93 |
| chr1_175157943_175158108 | 0.37 |
KIAA0040 |
KIAA0040 |
3865 |
0.32 |
| chr1_47133001_47133167 | 0.37 |
ATPAF1 |
ATP synthase mitochondrial F1 complex assembly factor 1 |
1015 |
0.38 |
| chr4_103266826_103266980 | 0.37 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
272 |
0.92 |
| chr10_104195058_104195350 | 0.36 |
ENSG00000202569 |
. |
1065 |
0.31 |
| chr14_95785313_95785520 | 0.36 |
CLMN |
calmin (calponin-like, transmembrane) |
827 |
0.72 |
| chr18_45526509_45526660 | 0.36 |
ZBTB7C |
zinc finger and BTB domain containing 7C |
40910 |
0.18 |
| chr10_104571858_104572081 | 0.35 |
ENSG00000252994 |
. |
8283 |
0.13 |
| chr14_69865476_69865674 | 0.34 |
SLC39A9 |
solute carrier family 39, member 9 |
166 |
0.59 |
| chrX_51636214_51636365 | 0.34 |
MAGED1 |
melanoma antigen family D, 1 |
340 |
0.91 |
| chr15_65592203_65592532 | 0.34 |
ENSG00000199568 |
. |
3978 |
0.15 |
| chr15_31652269_31652420 | 0.34 |
KLF13 |
Kruppel-like factor 13 |
6013 |
0.32 |
| chr13_74339708_74339938 | 0.33 |
KLF12 |
Kruppel-like factor 12 |
229363 |
0.02 |
| chr13_97874593_97875664 | 0.33 |
MBNL2 |
muscleblind-like splicing regulator 2 |
519 |
0.87 |
| chr11_62571959_62572195 | 0.32 |
NXF1 |
nuclear RNA export factor 1 |
420 |
0.56 |
| chr16_53986265_53986480 | 0.32 |
FTO |
fat mass and obesity associated |
18430 |
0.19 |
| chr1_32718292_32718517 | 0.32 |
LCK |
lymphocyte-specific protein tyrosine kinase |
1529 |
0.2 |
| chr16_18033135_18033286 | 0.31 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
384882 |
0.01 |
| chr12_48499913_48500348 | 0.31 |
SENP1 |
SUMO1/sentrin specific peptidase 1 |
39 |
0.49 |
| chr8_82024010_82024221 | 0.30 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
188 |
0.97 |
| chr20_54998016_54998238 | 0.30 |
CASS4 |
Cas scaffolding protein family member 4 |
10810 |
0.13 |
| chr9_133643618_133643769 | 0.30 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
54326 |
0.12 |
| chr13_49721188_49721557 | 0.30 |
FNDC3A |
fibronectin type III domain containing 3A |
36923 |
0.16 |
| chr18_43267753_43268033 | 0.29 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
21786 |
0.17 |
| chr14_52739518_52739669 | 0.29 |
PTGDR |
prostaglandin D2 receptor (DP) |
5063 |
0.28 |
| chr22_17567534_17567787 | 0.29 |
IL17RA |
interleukin 17 receptor A |
1811 |
0.31 |
| chr6_143250907_143251058 | 0.29 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
15356 |
0.25 |
| chr22_27070409_27070560 | 0.28 |
CRYBA4 |
crystallin, beta A4 |
52556 |
0.12 |
| chr11_85773706_85774153 | 0.28 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
5893 |
0.24 |
| chr12_49760663_49760953 | 0.28 |
SPATS2 |
spermatogenesis associated, serine-rich 2 |
63 |
0.96 |
| chr19_39728332_39728587 | 0.28 |
IFNL3 |
interferon, lambda 3 |
7187 |
0.12 |
| chr16_67517309_67517460 | 0.28 |
AGRP |
agouti related protein homolog (mouse) |
332 |
0.75 |
| chrY_1604612_1604993 | 0.27 |
NA |
NA |
> 106 |
NA |
| chr6_20206769_20207068 | 0.27 |
RP11-239H6.2 |
|
5400 |
0.25 |
| chr7_92875407_92875558 | 0.27 |
CCDC132 |
coiled-coil domain containing 132 |
13444 |
0.22 |
| chr17_73073685_73074098 | 0.27 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
9931 |
0.08 |
| chr19_3986525_3986905 | 0.27 |
EEF2 |
eukaryotic translation elongation factor 2 |
1248 |
0.29 |
| chr9_132098681_132099276 | 0.27 |
ENSG00000242281 |
. |
33762 |
0.17 |
| chr5_98369155_98369504 | 0.27 |
ENSG00000200351 |
. |
96878 |
0.08 |
| chr2_113033235_113033667 | 0.27 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
273 |
0.91 |
| chrX_1654663_1654996 | 0.27 |
P2RY8 |
purinergic receptor P2Y, G-protein coupled, 8 |
1171 |
0.53 |
| chr8_22398297_22398465 | 0.27 |
RP11-582J16.4 |
|
4537 |
0.13 |
| chr7_50348888_50349341 | 0.27 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
796 |
0.76 |
| chr8_38758238_38758438 | 0.27 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
415 |
0.84 |
| chr7_7297838_7297989 | 0.26 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
23962 |
0.22 |
| chr7_141437790_141437995 | 0.26 |
WEE2-AS1 |
WEE2 antisense RNA 1 |
128 |
0.63 |
| chr12_120967079_120967294 | 0.26 |
COQ5 |
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
201 |
0.89 |
| chr19_1028850_1029154 | 0.26 |
CNN2 |
calponin 2 |
2134 |
0.13 |
| chr15_101788492_101788643 | 0.26 |
CHSY1 |
chondroitin sulfate synthase 1 |
3570 |
0.21 |
| chr22_21771228_21771443 | 0.26 |
HIC2 |
hypermethylated in cancer 2 |
358 |
0.79 |
| chr16_56968887_56969454 | 0.26 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
144 |
0.93 |
| chr10_100227763_100227980 | 0.25 |
HPS1 |
Hermansky-Pudlak syndrome 1 |
21188 |
0.19 |
| chr7_55778225_55778376 | 0.25 |
FKBP9L |
FK506 binding protein 9-like |
2645 |
0.29 |
| chr1_36620951_36621452 | 0.25 |
MAP7D1 |
MAP7 domain containing 1 |
21 |
0.97 |
| chr7_50423750_50423901 | 0.25 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
56580 |
0.14 |
| chr9_123682878_123683088 | 0.25 |
TRAF1 |
TNF receptor-associated factor 1 |
6133 |
0.22 |
| chr7_44417747_44417898 | 0.25 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
52567 |
0.13 |
| chr8_8747685_8748237 | 0.25 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
3194 |
0.25 |
| chr1_36617295_36617817 | 0.25 |
TRAPPC3 |
trafficking protein particle complex 3 |
2458 |
0.21 |
| chr6_109802201_109802381 | 0.24 |
ZBTB24 |
zinc finger and BTB domain containing 24 |
2149 |
0.19 |
| chr2_113937983_113938134 | 0.24 |
AC016683.5 |
|
5121 |
0.15 |
| chr4_154418637_154418870 | 0.24 |
KIAA0922 |
KIAA0922 |
31252 |
0.2 |
| chr22_24109322_24109633 | 0.24 |
CHCHD10 |
coiled-coil-helix-coiled-coil-helix domain containing 10 |
586 |
0.54 |
| chr10_104956862_104957013 | 0.24 |
NT5C2 |
5'-nucleotidase, cytosolic II |
3881 |
0.25 |
| chr12_92518025_92518176 | 0.24 |
C12orf79 |
chromosome 12 open reading frame 79 |
12697 |
0.18 |
| chr15_86037970_86038121 | 0.24 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
49226 |
0.13 |
| chr5_148727679_148728000 | 0.24 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
2774 |
0.17 |
| chr7_127227981_127228310 | 0.23 |
ARF5 |
ADP-ribosylation factor 5 |
254 |
0.89 |
| chr10_99411142_99411293 | 0.23 |
PI4K2A |
phosphatidylinositol 4-kinase type 2 alpha |
10774 |
0.13 |
| chr2_74426850_74427050 | 0.23 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
1207 |
0.37 |
| chr11_82461800_82462168 | 0.23 |
FAM181B |
family with sequence similarity 181, member B |
17078 |
0.25 |
| chr20_52204433_52204907 | 0.23 |
ZNF217 |
zinc finger protein 217 |
5034 |
0.21 |
| chrX_9801459_9801610 | 0.23 |
ENSG00000206844 |
. |
41207 |
0.15 |
| chr1_155944536_155944687 | 0.23 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
2500 |
0.14 |
| chr10_71905035_71905382 | 0.23 |
TYSND1 |
trypsin domain containing 1 |
1134 |
0.47 |
| chr15_91137292_91137443 | 0.23 |
CRTC3 |
CREB regulated transcription coactivator 3 |
422 |
0.79 |
| chr1_9715239_9715390 | 0.22 |
C1orf200 |
chromosome 1 open reading frame 200 |
670 |
0.66 |
| chr6_16135670_16135821 | 0.22 |
ENSG00000263712 |
. |
6042 |
0.19 |
| chr12_114404344_114404564 | 0.22 |
RBM19 |
RNA binding motif protein 19 |
278 |
0.94 |
| chr11_62493191_62493431 | 0.22 |
HNRNPUL2 |
heterogeneous nuclear ribonucleoprotein U-like 2 |
1510 |
0.15 |
| chr9_132600041_132600192 | 0.22 |
USP20 |
ubiquitin specific peptidase 20 |
2359 |
0.21 |
| chr7_72721296_72721447 | 0.22 |
NSUN5 |
NOP2/Sun domain family, member 5 |
1430 |
0.32 |
| chr5_156569538_156569746 | 0.22 |
MED7 |
mediator complex subunit 7 |
144 |
0.5 |
| chr6_24949661_24949812 | 0.22 |
FAM65B |
family with sequence similarity 65, member B |
13548 |
0.22 |
| chr16_28890546_28890697 | 0.22 |
ATP2A1 |
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 |
57 |
0.87 |
| chr19_46018300_46018451 | 0.22 |
VASP |
vasodilator-stimulated phosphoprotein |
7626 |
0.11 |
| chr1_35659030_35659365 | 0.22 |
SFPQ |
splicing factor proline/glutamine-rich |
448 |
0.79 |
| chr20_39905047_39905444 | 0.22 |
ZHX3 |
zinc fingers and homeoboxes 3 |
4347 |
0.24 |
| chr11_65656225_65656433 | 0.22 |
FIBP |
fibroblast growth factor (acidic) intracellular binding protein |
319 |
0.71 |
| chr14_74416462_74416772 | 0.22 |
COQ6 |
coenzyme Q6 monooxygenase |
12 |
0.73 |
| chr13_21714248_21714458 | 0.22 |
ENSG00000252128 |
. |
257 |
0.52 |
| chr18_5295757_5296173 | 0.22 |
ZBTB14 |
zinc finger and BTB domain containing 14 |
74 |
0.98 |
| chr6_44041425_44041691 | 0.22 |
RP5-1120P11.1 |
|
831 |
0.6 |
| chr22_37822877_37823150 | 0.21 |
RP1-63G5.5 |
|
467 |
0.46 |
| chr11_575030_575208 | 0.21 |
PHRF1 |
PHD and ring finger domains 1 |
1367 |
0.21 |
| chrX_75371736_75371887 | 0.21 |
PBDC1 |
polysaccharide biosynthesis domain containing 1 |
20960 |
0.27 |
| chr6_152492731_152492882 | 0.21 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
3307 |
0.38 |
| chr2_11894079_11894417 | 0.21 |
LPIN1 |
lipin 1 |
7484 |
0.14 |
| chr3_46923397_46923551 | 0.21 |
MYL3 |
myosin, light chain 3, alkali; ventricular, skeletal, slow |
185 |
0.5 |
| chrX_75362891_75363042 | 0.21 |
PBDC1 |
polysaccharide biosynthesis domain containing 1 |
29805 |
0.25 |
| chr6_20570918_20571069 | 0.21 |
CDKAL1 |
CDK5 regulatory subunit associated protein 1-like 1 |
24421 |
0.21 |
| chr1_185663326_185663550 | 0.21 |
HMCN1 |
hemicentin 1 |
40245 |
0.17 |
| chr22_21872004_21872399 | 0.21 |
PI4KAP2 |
phosphatidylinositol 4-kinase, catalytic, alpha pseudogene 2 |
379 |
0.75 |
| chr15_94774265_94774529 | 0.21 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
370 |
0.93 |
| chr6_44899717_44900076 | 0.20 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
23351 |
0.28 |
| chr19_51873675_51874043 | 0.20 |
CLDND2 |
claudin domain containing 2 |
1602 |
0.15 |
| chr17_8026048_8026346 | 0.20 |
HES7 |
hes family bHLH transcription factor 7 |
1205 |
0.26 |
| chr8_42995208_42995412 | 0.20 |
HGSNAT |
heparan-alpha-glucosaminide N-acetyltransferase |
246 |
0.93 |
| chr9_123472752_123472978 | 0.20 |
MEGF9 |
multiple EGF-like-domains 9 |
3747 |
0.3 |
| chrY_1276930_1277081 | 0.20 |
NA |
NA |
> 106 |
NA |
| chr18_60824857_60825062 | 0.20 |
RP11-299P2.1 |
|
6406 |
0.25 |
| chr10_126406911_126407062 | 0.20 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
14233 |
0.17 |
| chr2_226329681_226329881 | 0.20 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
56184 |
0.18 |
| chrX_1326928_1327079 | 0.20 |
CRLF2 |
cytokine receptor-like factor 2 |
4524 |
0.21 |
| chr8_23397961_23398112 | 0.20 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
11463 |
0.16 |
| chr18_60823045_60823200 | 0.20 |
RP11-299P2.1 |
|
4569 |
0.27 |
| chrX_118369551_118369783 | 0.20 |
PGRMC1 |
progesterone receptor membrane component 1 |
549 |
0.83 |
| chr2_54786735_54787181 | 0.20 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
1427 |
0.42 |
| chr10_71929129_71929743 | 0.20 |
SAR1A |
SAR1 homolog A (S. cerevisiae) |
792 |
0.63 |
| chr17_75450125_75450790 | 0.20 |
SEPT9 |
septin 9 |
295 |
0.88 |
| chr1_155942988_155943139 | 0.20 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
952 |
0.36 |
| chr11_85956297_85957260 | 0.20 |
EED |
embryonic ectoderm development |
502 |
0.83 |
| chr1_31234118_31234269 | 0.20 |
LAPTM5 |
lysosomal protein transmembrane 5 |
3526 |
0.21 |
| chr12_52346063_52346405 | 0.20 |
ACVR1B |
activin A receptor, type IB |
706 |
0.62 |
| chrX_129220262_129220541 | 0.20 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
23935 |
0.18 |
| chr8_67626578_67626729 | 0.20 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
2000 |
0.3 |
| chr4_10459085_10459385 | 0.19 |
ZNF518B |
zinc finger protein 518B |
206 |
0.96 |
| chr18_13438227_13438398 | 0.19 |
LDLRAD4-AS1 |
LDLRAD4 antisense RNA 1 |
10833 |
0.12 |
| chr1_2163627_2164198 | 0.19 |
SKI |
v-ski avian sarcoma viral oncogene homolog |
3778 |
0.16 |
| chr6_43337603_43337930 | 0.19 |
ZNF318 |
zinc finger protein 318 |
585 |
0.7 |
| chr10_29096673_29096824 | 0.19 |
C10orf126 |
chromosome 10 open reading frame 126 |
38589 |
0.16 |
| chr17_66233779_66233987 | 0.19 |
AMZ2 |
archaelysin family metallopeptidase 2 |
9832 |
0.16 |
| chr9_104160646_104160869 | 0.19 |
MRPL50 |
mitochondrial ribosomal protein L50 |
129 |
0.79 |
| chr15_50644840_50645537 | 0.19 |
GABPB1 |
GA binding protein transcription factor, beta subunit 1 |
2133 |
0.21 |
| chr3_197353837_197354109 | 0.19 |
AC024560.3 |
|
746 |
0.69 |
| chr5_174905979_174906366 | 0.19 |
SFXN1 |
sideroflexin 1 |
577 |
0.78 |
| chr1_160548119_160548396 | 0.19 |
CD84 |
CD84 molecule |
1006 |
0.48 |
| chr1_235805657_235805845 | 0.19 |
GNG4 |
guanine nucleotide binding protein (G protein), gamma 4 |
7542 |
0.28 |
| chr14_75800906_75801057 | 0.19 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
54085 |
0.1 |
| chr2_97428078_97428820 | 0.19 |
CNNM4 |
cyclin M4 |
1810 |
0.3 |
| chr17_38490189_38490340 | 0.19 |
RARA |
retinoic acid receptor, alpha |
7376 |
0.11 |
| chr10_35775018_35775169 | 0.19 |
GJD4 |
gap junction protein, delta 4, 40.1kDa |
119245 |
0.05 |
| chr10_73847731_73848399 | 0.19 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
21 |
0.98 |
| chr19_5624316_5624482 | 0.19 |
SAFB2 |
scaffold attachment factor B2 |
342 |
0.77 |
| chr17_57230990_57231141 | 0.19 |
SKA2 |
spindle and kinetochore associated complex subunit 2 |
1470 |
0.21 |
| chr17_74731558_74731709 | 0.19 |
MFSD11 |
major facilitator superfamily domain containing 11 |
314 |
0.67 |
| chr1_12139606_12139757 | 0.19 |
ENSG00000201135 |
. |
1640 |
0.3 |
| chr14_39899307_39899586 | 0.18 |
FBXO33 |
F-box protein 33 |
1633 |
0.47 |
| chr10_102026913_102027138 | 0.18 |
CWF19L1 |
CWF19-like 1, cell cycle control (S. pombe) |
412 |
0.75 |
| chr19_34663821_34664709 | 0.18 |
LSM14A |
LSM14A, SCD6 homolog A (S. cerevisiae) |
702 |
0.77 |
| chr12_57030254_57030540 | 0.18 |
BAZ2A |
bromodomain adjacent to zinc finger domain, 2A |
234 |
0.88 |
| chr13_47242269_47242651 | 0.18 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
11511 |
0.28 |
| chr20_52259090_52259799 | 0.18 |
ENSG00000238468 |
. |
25853 |
0.18 |
| chrY_15017679_15018174 | 0.18 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
277 |
0.95 |
| chr13_99952403_99952624 | 0.18 |
GPR183 |
G protein-coupled receptor 183 |
7146 |
0.21 |
| chr7_90339651_90339847 | 0.18 |
CDK14 |
cyclin-dependent kinase 14 |
573 |
0.85 |
| chr1_32665798_32666702 | 0.18 |
CCDC28B |
coiled-coil domain containing 28B |
16 |
0.94 |
| chr1_209921149_209921488 | 0.18 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
8059 |
0.14 |
| chr11_65342200_65342895 | 0.18 |
EHBP1L1 |
EH domain binding protein 1-like 1 |
962 |
0.29 |
| chr22_20428028_20428487 | 0.18 |
ENSG00000271796 |
. |
5896 |
0.13 |
| chr1_21503574_21503811 | 0.18 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
315 |
0.69 |
| chr18_24867919_24868070 | 0.18 |
CHST9 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
102713 |
0.09 |
| chr1_165600518_165600732 | 0.18 |
MGST3 |
microsomal glutathione S-transferase 3 |
133 |
0.96 |
| chr11_133797734_133798200 | 0.18 |
IGSF9B |
immunoglobulin superfamily, member 9B |
18220 |
0.18 |
| chr11_62308687_62309106 | 0.18 |
RP11-864I4.3 |
|
4112 |
0.1 |
| chr2_29803342_29803493 | 0.18 |
ENSG00000266310 |
. |
76558 |
0.11 |
| chr1_246280752_246281030 | 0.18 |
RP11-36N20.1 |
|
8963 |
0.25 |
| chr7_38389675_38390189 | 0.18 |
AMPH |
amphiphysin |
112781 |
0.07 |
| chr2_106362063_106362974 | 0.17 |
NCK2 |
NCK adaptor protein 2 |
330 |
0.94 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.4 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.1 | 0.2 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.2 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.4 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.3 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation involved in immune response(GO:0002313) mature B cell differentiation(GO:0002335) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.6 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.2 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 2.1 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.1 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) |
| 0.0 | 0.0 | GO:0045588 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.1 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.1 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0051132 | NK T cell activation(GO:0051132) |
| 0.0 | 0.1 | GO:0046015 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0051000 | positive regulation of nitric-oxide synthase activity(GO:0051000) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.0 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0046471 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.0 | GO:0002707 | negative regulation of lymphocyte mediated immunity(GO:0002707) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0042921 | glucocorticoid receptor signaling pathway(GO:0042921) |
| 0.0 | 0.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.0 | GO:1903504 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.0 | GO:0070431 | cytoplasmic pattern recognition receptor signaling pathway(GO:0002753) nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway(GO:0035872) nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.0 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.0 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0071599 | otic vesicle formation(GO:0030916) otic vesicle development(GO:0071599) otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.3 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.0 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0030288 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.0 | GO:0070061 | fructose binding(GO:0070061) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.2 | GO:0004428 | obsolete inositol or phosphatidylinositol kinase activity(GO:0004428) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.0 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.1 | 1.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |