Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
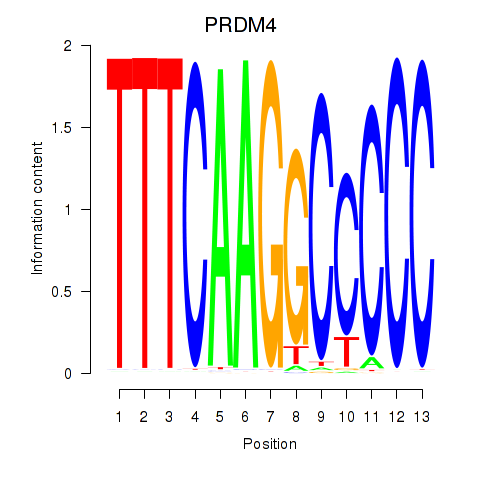
Results for PRDM4
Z-value: 0.80
Transcription factors associated with PRDM4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PRDM4
|
ENSG00000110851.7 | PR/SET domain 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_108143303_108143479 | PRDM4 | 2161 | 0.269624 | -0.88 | 1.8e-03 | Click! |
| chr12_108142710_108142874 | PRDM4 | 2760 | 0.230268 | -0.65 | 5.9e-02 | Click! |
| chr12_108153036_108153205 | PRDM4 | 1585 | 0.344352 | -0.53 | 1.4e-01 | Click! |
| chr12_108155132_108155624 | PRDM4 | 441 | 0.804266 | 0.36 | 3.4e-01 | Click! |
| chr12_108153552_108153777 | PRDM4 | 1041 | 0.489436 | -0.32 | 4.0e-01 | Click! |
Activity of the PRDM4 motif across conditions
Conditions sorted by the z-value of the PRDM4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
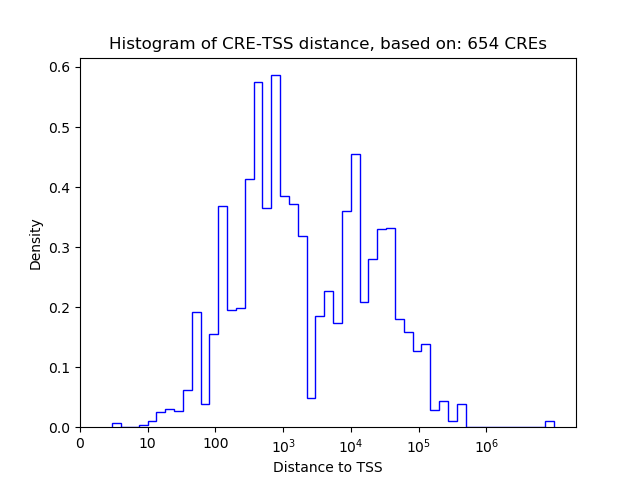
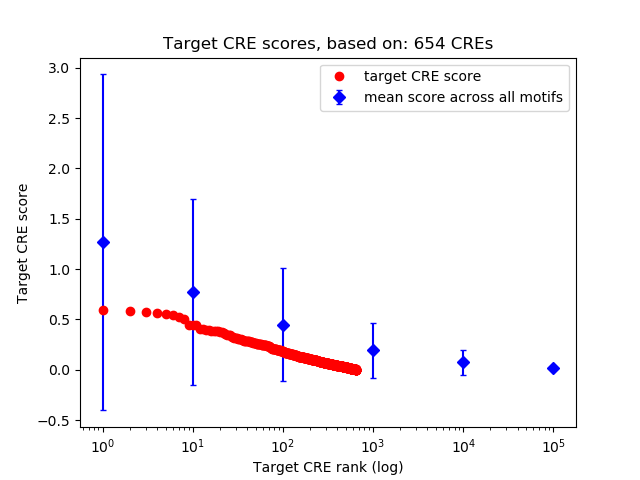
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_158274562_158274945 | 0.60 |
CYTIP |
cytohesin 1 interacting protein |
21173 |
0.2 |
| chr6_32159695_32160229 | 0.58 |
GPSM3 |
G-protein signaling modulator 3 |
683 |
0.42 |
| chr19_50003533_50003936 | 0.57 |
hsa-mir-150 |
hsa-mir-150 |
47 |
0.8 |
| chr1_9699551_9699702 | 0.56 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
12164 |
0.16 |
| chr15_33383427_33383578 | 0.56 |
FMN1 |
formin 1 |
23139 |
0.25 |
| chrX_153191913_153192277 | 0.54 |
ARHGAP4 |
Rho GTPase activating protein 4 |
116 |
0.92 |
| chr10_121276429_121276711 | 0.53 |
RGS10 |
regulator of G-protein signaling 10 |
10419 |
0.24 |
| chr14_22771842_22772128 | 0.51 |
ENSG00000251002 |
. |
129734 |
0.04 |
| chr19_11640031_11640182 | 0.44 |
ECSIT |
ECSIT signalling integrator |
117 |
0.92 |
| chr6_15253008_15253181 | 0.44 |
JARID2 |
jumonji, AT rich interactive domain 2 |
3951 |
0.23 |
| chr7_38389357_38389573 | 0.44 |
AMPH |
amphiphysin |
113248 |
0.07 |
| chr7_147856085_147856236 | 0.41 |
CNTNAP2 |
contactin associated protein-like 2 |
25384 |
0.26 |
| chr14_105531964_105532391 | 0.41 |
GPR132 |
G protein-coupled receptor 132 |
395 |
0.85 |
| chr9_107591981_107592132 | 0.40 |
NIPSNAP3B |
nipsnap homolog 3B (C. elegans) |
65605 |
0.11 |
| chr1_111742376_111742661 | 0.39 |
DENND2D |
DENN/MADD domain containing 2D |
793 |
0.38 |
| chrX_9431281_9431551 | 0.39 |
TBL1X |
transducin (beta)-like 1X-linked |
45 |
0.99 |
| chr7_90893538_90893822 | 0.39 |
FZD1 |
frizzled family receptor 1 |
103 |
0.99 |
| chr3_170963778_170963929 | 0.38 |
TNIK |
TRAF2 and NCK interacting kinase |
20353 |
0.26 |
| chr5_1315569_1315720 | 0.38 |
ENSG00000263670 |
. |
6152 |
0.18 |
| chr14_23010806_23011164 | 0.38 |
TRAJ15 |
T cell receptor alpha joining 15 |
12405 |
0.1 |
| chr2_231788728_231788879 | 0.37 |
GPR55 |
G protein-coupled receptor 55 |
1138 |
0.46 |
| chr1_198622996_198623151 | 0.37 |
RP11-553K8.5 |
|
13117 |
0.23 |
| chr1_159037657_159037949 | 0.36 |
ENSG00000265589 |
. |
8764 |
0.18 |
| chr14_100537247_100537438 | 0.35 |
CTD-2376I20.1 |
|
3873 |
0.18 |
| chr10_88170804_88170968 | 0.34 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
44651 |
0.14 |
| chr15_101782791_101782964 | 0.34 |
CHSY1 |
chondroitin sulfate synthase 1 |
9260 |
0.18 |
| chrX_48776392_48776920 | 0.33 |
PIM2 |
pim-2 oncogene |
355 |
0.76 |
| chr1_226066769_226066920 | 0.33 |
TMEM63A |
transmembrane protein 63A |
1465 |
0.3 |
| chr2_10169450_10169738 | 0.32 |
KLF11 |
Kruppel-like factor 11 |
13382 |
0.13 |
| chr22_31625511_31626031 | 0.31 |
ENSG00000202019 |
. |
387 |
0.76 |
| chr6_41169596_41169747 | 0.31 |
TREML2 |
triggering receptor expressed on myeloid cells-like 2 |
739 |
0.55 |
| chr16_21519015_21519166 | 0.31 |
ENSG00000265462 |
. |
1634 |
0.33 |
| chr22_50050359_50050996 | 0.31 |
C22orf34 |
chromosome 22 open reading frame 34 |
401 |
0.87 |
| chr10_112287695_112287846 | 0.30 |
DUSP5 |
dual specificity phosphatase 5 |
30174 |
0.14 |
| chr12_125477524_125477679 | 0.30 |
BRI3BP |
BRI3 binding protein |
645 |
0.71 |
| chr11_441689_441900 | 0.29 |
ANO9 |
anoctamin 9 |
217 |
0.82 |
| chr2_12862265_12862722 | 0.29 |
TRIB2 |
tribbles pseudokinase 2 |
4129 |
0.3 |
| chr5_17258045_17258196 | 0.29 |
ENSG00000252908 |
. |
17400 |
0.19 |
| chr2_234296852_234297003 | 0.29 |
DGKD |
diacylglycerol kinase, delta 130kDa |
127 |
0.96 |
| chr9_99175747_99176261 | 0.29 |
ZNF367 |
zinc finger protein 367 |
4607 |
0.23 |
| chr3_16884765_16884916 | 0.29 |
PLCL2 |
phospholipase C-like 2 |
41612 |
0.19 |
| chr11_127910618_127910769 | 0.28 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
464596 |
0.01 |
| chr6_126067758_126068417 | 0.28 |
HEY2 |
hes-related family bHLH transcription factor with YRPW motif 2 |
723 |
0.57 |
| chr8_103136777_103137074 | 0.28 |
NCALD |
neurocalcin delta |
117 |
0.91 |
| chr17_80806215_80806449 | 0.28 |
ZNF750 |
zinc finger protein 750 |
7878 |
0.16 |
| chr1_147013251_147013702 | 0.27 |
BCL9 |
B-cell CLL/lymphoma 9 |
294 |
0.94 |
| chr2_146013147_146013298 | 0.27 |
ENSG00000253036 |
. |
79416 |
0.12 |
| chr17_76254452_76254830 | 0.27 |
TMEM235 |
transmembrane protein 235 |
26519 |
0.12 |
| chrX_147583270_147583557 | 0.27 |
AFF2 |
AF4/FMR2 family, member 2 |
884 |
0.51 |
| chrX_30849195_30849346 | 0.26 |
TAB3-AS1 |
TAB3 antisense RNA 1 |
3470 |
0.24 |
| chr6_32408572_32408723 | 0.26 |
HLA-DRA |
major histocompatibility complex, class II, DR alpha |
992 |
0.48 |
| chr2_9613939_9614127 | 0.26 |
IAH1 |
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) |
86 |
0.97 |
| chr5_34916244_34916587 | 0.26 |
RAD1 |
RAD1 homolog (S. pombe) |
456 |
0.67 |
| chr7_101784250_101784577 | 0.25 |
ENSG00000252824 |
. |
53031 |
0.12 |
| chr1_172502667_172502917 | 0.25 |
SUCO |
SUN domain containing ossification factor |
435 |
0.87 |
| chr17_33640471_33640814 | 0.25 |
SLFN11 |
schlafen family member 11 |
39508 |
0.12 |
| chr6_42898071_42898286 | 0.25 |
CNPY3 |
canopy FGF signaling regulator 3 |
1196 |
0.31 |
| chr1_193153819_193153970 | 0.25 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
1890 |
0.36 |
| chr9_77762612_77762996 | 0.25 |
ENSG00000200041 |
. |
33733 |
0.15 |
| chr4_107236640_107237312 | 0.25 |
AIMP1 |
aminoacyl tRNA synthetase complex-interacting multifunctional protein 1 |
123 |
0.8 |
| chr1_84902979_84903163 | 0.24 |
DNASE2B |
deoxyribonuclease II beta |
29158 |
0.16 |
| chr11_66887899_66888432 | 0.24 |
KDM2A |
lysine (K)-specific demethylase 2A |
1007 |
0.52 |
| chr2_26101893_26102047 | 0.24 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
585 |
0.82 |
| chr16_31548282_31548433 | 0.24 |
AHSP |
alpha hemoglobin stabilizing protein |
9059 |
0.11 |
| chr7_157306639_157306790 | 0.24 |
AC006372.6 |
|
726 |
0.7 |
| chr2_43438121_43438285 | 0.24 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
15545 |
0.22 |
| chr14_103011281_103011526 | 0.24 |
ENSG00000266015 |
. |
5422 |
0.18 |
| chr14_32030134_32030320 | 0.24 |
CTD-2213F21.3 |
|
313 |
0.52 |
| chr21_39844669_39844875 | 0.24 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
25573 |
0.26 |
| chr12_58132957_58133143 | 0.23 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
1021 |
0.26 |
| chr15_74286545_74286747 | 0.23 |
STOML1 |
stomatin (EPB72)-like 1 |
317 |
0.52 |
| chr9_134145569_134145886 | 0.23 |
FAM78A |
family with sequence similarity 78, member A |
153 |
0.95 |
| chr7_5729023_5729311 | 0.22 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
9075 |
0.21 |
| chr1_27114087_27114296 | 0.22 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
228 |
0.89 |
| chr17_32573755_32573906 | 0.21 |
CCL2 |
chemokine (C-C motif) ligand 2 |
8474 |
0.13 |
| chr8_67524543_67525469 | 0.21 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
136 |
0.96 |
| chr17_73008873_73009557 | 0.21 |
ICT1 |
immature colon carcinoma transcript 1 |
446 |
0.66 |
| chr3_107818747_107818903 | 0.21 |
CD47 |
CD47 molecule |
8953 |
0.3 |
| chr4_124542421_124542572 | 0.21 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
221373 |
0.02 |
| chr16_68104203_68104354 | 0.21 |
ENSG00000221789 |
. |
14491 |
0.09 |
| chr1_3446831_3447137 | 0.21 |
MEGF6 |
multiple EGF-like-domains 6 |
1028 |
0.53 |
| chr3_171758588_171759081 | 0.21 |
FNDC3B |
fibronectin type III domain containing 3B |
489 |
0.88 |
| chr3_133291490_133292033 | 0.20 |
CDV3 |
CDV3 homolog (mouse) |
813 |
0.65 |
| chrX_48544144_48544295 | 0.20 |
WAS |
Wiskott-Aldrich syndrome |
2051 |
0.22 |
| chr17_42634478_42634800 | 0.20 |
FZD2 |
frizzled family receptor 2 |
286 |
0.91 |
| chr17_75178693_75178860 | 0.20 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
1657 |
0.41 |
| chr12_27038132_27038283 | 0.20 |
ASUN |
asunder spermatogenesis regulator |
32460 |
0.15 |
| chr3_56817061_56817212 | 0.20 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
7451 |
0.29 |
| chr18_12703898_12704049 | 0.20 |
PSMG2 |
proteasome (prosome, macropain) assembly chaperone 2 |
951 |
0.39 |
| chr22_40743199_40743350 | 0.20 |
ADSL |
adenylosuccinate lyase |
738 |
0.66 |
| chr13_99740074_99740470 | 0.19 |
DOCK9-AS2 |
DOCK9 antisense RNA 2 (head to head) |
199 |
0.92 |
| chrX_77359847_77360362 | 0.19 |
PGK1 |
phosphoglycerate kinase 1 |
352 |
0.91 |
| chr14_64010347_64011034 | 0.19 |
CTD-2302E22.4 |
|
403 |
0.59 |
| chr5_90478735_90478954 | 0.19 |
ENSG00000199643 |
. |
87700 |
0.1 |
| chr12_29310650_29310817 | 0.19 |
FAR2 |
fatty acyl CoA reductase 2 |
8600 |
0.26 |
| chr19_54881402_54882266 | 0.19 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
224 |
0.88 |
| chr1_32293954_32294105 | 0.18 |
SPOCD1 |
SPOC domain containing 1 |
12377 |
0.16 |
| chr10_30990441_30990915 | 0.18 |
SVILP1 |
supervillin pseudogene 1 |
5828 |
0.28 |
| chrX_24482692_24483178 | 0.18 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
403 |
0.88 |
| chr7_30068476_30069002 | 0.18 |
PLEKHA8 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
438 |
0.77 |
| chr9_139559538_139560114 | 0.18 |
EGFL7 |
EGF-like-domain, multiple 7 |
418 |
0.68 |
| chr19_14358666_14358817 | 0.18 |
ENSG00000240803 |
. |
34877 |
0.12 |
| chr10_63809082_63810748 | 0.18 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
945 |
0.69 |
| chr17_28995985_28996136 | 0.18 |
ENSG00000241631 |
. |
33733 |
0.12 |
| chr15_75082012_75082172 | 0.17 |
ENSG00000264386 |
. |
994 |
0.33 |
| chr3_169381606_169381757 | 0.17 |
MECOM |
MDS1 and EVI1 complex locus |
423 |
0.83 |
| chr19_46172257_46172467 | 0.17 |
GIPR |
gastric inhibitory polypeptide receptor |
847 |
0.37 |
| chr20_35823464_35823615 | 0.17 |
MROH8 |
maestro heat-like repeat family member 8 |
15548 |
0.18 |
| chr1_184794791_184794942 | 0.17 |
ENSG00000252222 |
. |
4243 |
0.26 |
| chr19_55009541_55009776 | 0.17 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
558 |
0.61 |
| chr7_103847754_103848404 | 0.17 |
ORC5 |
origin recognition complex, subunit 5 |
326 |
0.94 |
| chr1_28563316_28563564 | 0.17 |
ATPIF1 |
ATPase inhibitory factor 1 |
775 |
0.48 |
| chr15_56335461_56335612 | 0.17 |
ENSG00000239703 |
. |
13357 |
0.23 |
| chr9_86593584_86594514 | 0.16 |
HNRNPK |
heterogeneous nuclear ribonucleoprotein K |
682 |
0.51 |
| chr5_72634404_72634555 | 0.16 |
FOXD1 |
forkhead box D1 |
109873 |
0.06 |
| chr7_156400256_156401034 | 0.16 |
ENSG00000182648 |
. |
2187 |
0.32 |
| chr19_926201_926965 | 0.16 |
ARID3A |
AT rich interactive domain 3A (BRIGHT-like) |
583 |
0.51 |
| chr12_128850300_128850451 | 0.16 |
ENSG00000238895 |
. |
32602 |
0.2 |
| chr12_92534627_92535592 | 0.16 |
C12orf79 |
chromosome 12 open reading frame 79 |
1335 |
0.39 |
| chr17_7741127_7741463 | 0.16 |
KDM6B |
lysine (K)-specific demethylase 6B |
1927 |
0.19 |
| chr3_53288573_53288749 | 0.16 |
TKT |
transketolase |
1355 |
0.36 |
| chr8_103801561_103801807 | 0.16 |
ENSG00000266799 |
. |
55737 |
0.11 |
| chrX_48937651_48938132 | 0.16 |
WDR45 |
WD repeat domain 45 |
24 |
0.81 |
| chrX_75393364_75393724 | 0.15 |
PBDC1 |
polysaccharide biosynthesis domain containing 1 |
669 |
0.81 |
| chr10_29771962_29772274 | 0.15 |
SVIL |
supervillin |
13329 |
0.23 |
| chr19_52553655_52553806 | 0.15 |
ZNF432 |
zinc finger protein 432 |
1657 |
0.24 |
| chr15_98511098_98511399 | 0.15 |
ARRDC4 |
arrestin domain containing 4 |
7320 |
0.25 |
| chr1_46712530_46712975 | 0.15 |
RAD54L |
RAD54-like (S. cerevisiae) |
615 |
0.66 |
| chr13_30688522_30688966 | 0.15 |
ENSG00000266816 |
. |
92092 |
0.09 |
| chrX_117767379_117767530 | 0.15 |
ENSG00000206862 |
. |
63976 |
0.12 |
| chr14_70161155_70161306 | 0.15 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
32387 |
0.19 |
| chr17_41324442_41324699 | 0.15 |
NBR1 |
neighbor of BRCA1 gene 1 |
1310 |
0.36 |
| chr5_179335042_179335255 | 0.15 |
TBC1D9B |
TBC1 domain family, member 9B (with GRAM domain) |
289 |
0.88 |
| chr5_100236594_100237334 | 0.15 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
1954 |
0.48 |
| chr6_6724392_6724959 | 0.15 |
LY86-AS1 |
LY86 antisense RNA 1 |
101671 |
0.08 |
| chr1_43232974_43233556 | 0.15 |
C1orf50 |
chromosome 1 open reading frame 50 |
288 |
0.65 |
| chr7_961818_962008 | 0.14 |
ADAP1 |
ArfGAP with dual PH domains 1 |
1387 |
0.35 |
| chr3_37156794_37156945 | 0.14 |
ENSG00000206645 |
. |
22144 |
0.14 |
| chr1_231748956_231749347 | 0.14 |
LINC00582 |
long intergenic non-protein coding RNA 582 |
1315 |
0.46 |
| chrX_56814440_56814591 | 0.14 |
ENSG00000204272 |
. |
58823 |
0.16 |
| chrX_64888680_64889016 | 0.14 |
MSN |
moesin |
1311 |
0.61 |
| chr2_176237854_176238005 | 0.14 |
ENSG00000221347 |
. |
42828 |
0.19 |
| chr17_5186908_5187121 | 0.14 |
RABEP1 |
rabaptin, RAB GTPase binding effector protein 1 |
1232 |
0.43 |
| chr12_97300286_97300520 | 0.14 |
NEDD1 |
neural precursor cell expressed, developmentally down-regulated 1 |
598 |
0.84 |
| chr7_32535632_32536288 | 0.14 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
638 |
0.62 |
| chr6_113201994_113202335 | 0.14 |
ENSG00000201386 |
. |
90531 |
0.1 |
| chr16_77756424_77756870 | 0.13 |
NUDT7 |
nudix (nucleoside diphosphate linked moiety X)-type motif 7 |
205 |
0.94 |
| chr4_122722562_122723305 | 0.13 |
EXOSC9 |
exosome component 9 |
185 |
0.94 |
| chr2_239800244_239800395 | 0.13 |
TWIST2 |
twist family bHLH transcription factor 2 |
43646 |
0.15 |
| chr12_113494281_113494869 | 0.13 |
DTX1 |
deltex homolog 1 (Drosophila) |
920 |
0.55 |
| chr15_57853592_57853743 | 0.13 |
GCOM1 |
GRINL1A complex locus 1 |
30439 |
0.16 |
| chr12_58240459_58240718 | 0.13 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
66 |
0.95 |
| chr10_75634514_75635168 | 0.13 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
498 |
0.7 |
| chr12_4380598_4381153 | 0.13 |
CCND2 |
cyclin D2 |
2063 |
0.27 |
| chr5_168278845_168278996 | 0.13 |
ENSG00000202345 |
. |
60417 |
0.13 |
| chrX_69354363_69354644 | 0.13 |
IGBP1 |
immunoglobulin (CD79A) binding protein 1 |
1190 |
0.39 |
| chr8_21776103_21776686 | 0.13 |
XPO7 |
exportin 7 |
786 |
0.64 |
| chr2_204192506_204192763 | 0.13 |
ABI2 |
abl-interactor 2 |
308 |
0.86 |
| chr1_145516788_145517229 | 0.13 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
448 |
0.67 |
| chr9_93569627_93569778 | 0.13 |
SYK |
spleen tyrosine kinase |
5493 |
0.35 |
| chr8_57423628_57423790 | 0.12 |
RP11-17A4.2 |
|
22052 |
0.24 |
| chr6_89791603_89792706 | 0.12 |
PNRC1 |
proline-rich nuclear receptor coactivator 1 |
599 |
0.61 |
| chr12_69979635_69980167 | 0.12 |
CCT2 |
chaperonin containing TCP1, subunit 2 (beta) |
455 |
0.72 |
| chr3_63956535_63956875 | 0.12 |
ATXN7 |
ataxin 7 |
3285 |
0.2 |
| chr11_10480387_10480598 | 0.12 |
AMPD3 |
adenosine monophosphate deaminase 3 |
2195 |
0.31 |
| chr9_98078977_98079489 | 0.12 |
FANCC |
Fanconi anemia, complementation group C |
57 |
0.98 |
| chr11_72436543_72436694 | 0.12 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
1555 |
0.28 |
| chr11_66792086_66792237 | 0.12 |
SYT12 |
synaptotagmin XII |
1345 |
0.36 |
| chr1_171750817_171751440 | 0.12 |
METTL13 |
methyltransferase like 13 |
286 |
0.89 |
| chr5_133860075_133861084 | 0.12 |
JADE2 |
jade family PHD finger 2 |
576 |
0.71 |
| chr10_81204131_81204808 | 0.12 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
494 |
0.82 |
| chr4_83719416_83720536 | 0.12 |
SCD5 |
stearoyl-CoA desaturase 5 |
34 |
0.98 |
| chr19_44281250_44281441 | 0.12 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
4064 |
0.15 |
| chr21_35445889_35446081 | 0.12 |
SLC5A3 |
solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
115 |
0.57 |
| chr19_16250491_16250727 | 0.12 |
CTD-2231E14.8 |
|
22 |
0.96 |
| chr17_59530124_59530284 | 0.12 |
TBX4 |
T-box 4 |
425 |
0.8 |
| chr17_56709506_56709776 | 0.12 |
ENSG00000212195 |
. |
444 |
0.75 |
| chr1_198904942_198905093 | 0.12 |
ENSG00000207759 |
. |
76735 |
0.11 |
| chr10_94608596_94609267 | 0.12 |
EXOC6 |
exocyst complex component 6 |
654 |
0.81 |
| chr6_4135067_4135363 | 0.12 |
RP3-400B16.4 |
|
442 |
0.49 |
| chr4_185570944_185571157 | 0.12 |
PRIMPOL |
primase and polymerase (DNA-directed) |
171 |
0.75 |
| chr16_87985408_87986210 | 0.12 |
BANP |
BTG3 associated nuclear protein |
716 |
0.69 |
| chrX_47054421_47054572 | 0.12 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
1255 |
0.34 |
| chr5_134879041_134879292 | 0.12 |
NEUROG1 |
neurogenin 1 |
7527 |
0.18 |
| chr19_6108632_6109202 | 0.11 |
CTB-66B24.1 |
|
416 |
0.75 |
| chr19_16296249_16296834 | 0.11 |
FAM32A |
family with sequence similarity 32, member A |
275 |
0.85 |
| chr2_12859088_12859514 | 0.11 |
TRIB2 |
tribbles pseudokinase 2 |
937 |
0.67 |
| chr16_57168681_57168893 | 0.11 |
CPNE2 |
copine II |
15676 |
0.14 |
| chr4_111532497_111532648 | 0.11 |
PITX2 |
paired-like homeodomain 2 |
11635 |
0.27 |
| chr5_131826766_131827319 | 0.11 |
IRF1 |
interferon regulatory factor 1 |
552 |
0.69 |
| chr11_61185467_61185618 | 0.11 |
CPSF7 |
cleavage and polyadenylation specific factor 7, 59kDa |
11234 |
0.1 |
| chr20_46377193_46377344 | 0.11 |
SULF2 |
sulfatase 2 |
36965 |
0.17 |
| chr6_149082246_149082558 | 0.11 |
UST |
uronyl-2-sulfotransferase |
13938 |
0.3 |
| chr1_103731186_103731553 | 0.11 |
COL11A1 |
collagen, type XI, alpha 1 |
157317 |
0.04 |
| chr19_36398950_36399155 | 0.11 |
TYROBP |
TYRO protein tyrosine kinase binding protein |
97 |
0.93 |
| chr10_82260474_82260649 | 0.11 |
RP11-137H2.4 |
|
35137 |
0.15 |
| chr2_33174000_33174151 | 0.11 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
1706 |
0.38 |
| chr7_100450418_100451115 | 0.11 |
SLC12A9 |
solute carrier family 12, member 9 |
142 |
0.81 |
| chr19_872645_872938 | 0.11 |
CFD |
complement factor D (adipsin) |
13101 |
0.07 |
| chr17_45331851_45332126 | 0.11 |
ITGB3 |
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
725 |
0.57 |
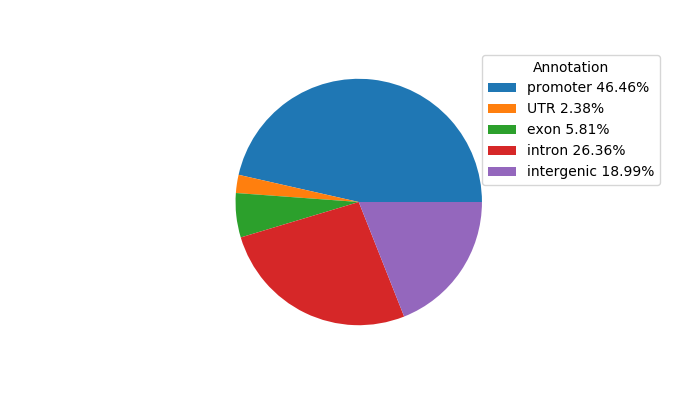
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.6 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.1 | 0.2 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.2 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0045714 | regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045714) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.0 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.2 | GO:0007128 | meiotic prophase I(GO:0007128) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.4 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.0 | 0.1 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.7 | GO:0032589 | neuron projection membrane(GO:0032589) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |