Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
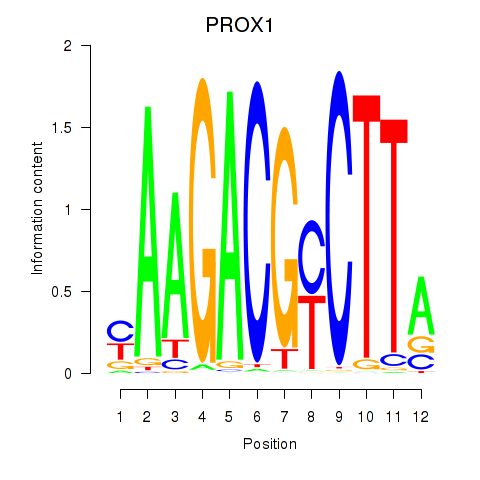
Results for PROX1
Z-value: 1.72
Transcription factors associated with PROX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PROX1
|
ENSG00000117707.11 | prospero homeobox 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_214170628_214170779 | PROX1 | 824 | 0.660475 | 0.45 | 2.2e-01 | Click! |
| chr1_214156478_214156705 | PROX1 | 67 | 0.975804 | -0.42 | 2.7e-01 | Click! |
| chr1_214161166_214161486 | PROX1 | 40 | 0.845613 | 0.24 | 5.4e-01 | Click! |
| chr1_214170321_214170472 | PROX1 | 517 | 0.812497 | 0.24 | 5.4e-01 | Click! |
| chr1_214156195_214156346 | PROX1 | 254 | 0.928582 | -0.16 | 6.9e-01 | Click! |
Activity of the PROX1 motif across conditions
Conditions sorted by the z-value of the PROX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
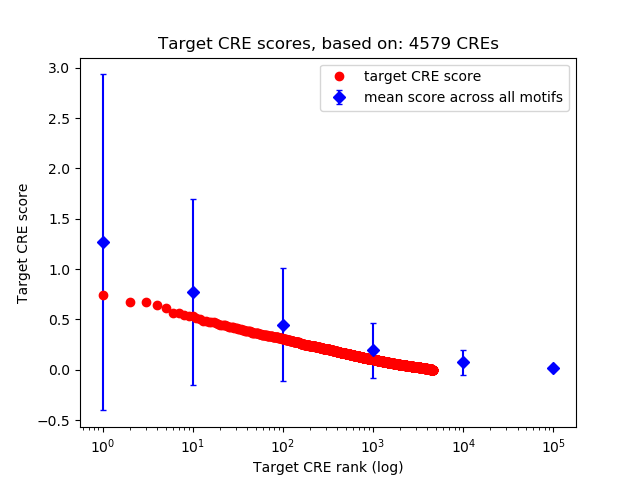
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_8835423_8835602 | 0.74 |
ENSG00000201348 |
. |
23679 |
0.24 |
| chr2_108443697_108444171 | 0.68 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
541 |
0.87 |
| chr6_42879136_42879323 | 0.67 |
PTCRA |
pre T-cell antigen receptor alpha |
4498 |
0.12 |
| chr2_108445323_108445660 | 0.65 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
2098 |
0.48 |
| chr12_49503707_49503930 | 0.62 |
LMBR1L |
limb development membrane protein 1-like |
170 |
0.89 |
| chr14_63671686_63671939 | 0.57 |
RHOJ |
ras homolog family member J |
235 |
0.95 |
| chr2_101179059_101179268 | 0.57 |
PDCL3 |
phosducin-like 3 |
11 |
0.97 |
| chr9_98412049_98412205 | 0.54 |
DKFZP434H0512 |
Protein LOC100506667; Putative uncharacterized protein DKFZp434H0512 |
122478 |
0.05 |
| chr5_88028176_88028469 | 0.54 |
CTC-467M3.2 |
|
39854 |
0.16 |
| chr1_171454288_171454467 | 0.53 |
PRRC2C |
proline-rich coiled-coil 2C |
274 |
0.88 |
| chr1_120685462_120685613 | 0.52 |
ENSG00000207149 |
. |
15975 |
0.24 |
| chr14_64663537_64663695 | 0.51 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
5295 |
0.27 |
| chr14_33194390_33194567 | 0.49 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
10579 |
0.32 |
| chr3_112356800_112357010 | 0.48 |
CCDC80 |
coiled-coil domain containing 80 |
39 |
0.98 |
| chr19_16188890_16189584 | 0.48 |
TPM4 |
tropomyosin 4 |
1406 |
0.39 |
| chr10_70847938_70848765 | 0.48 |
SRGN |
serglycin |
477 |
0.82 |
| chrX_153095524_153095861 | 0.48 |
PDZD4 |
PDZ domain containing 4 |
121 |
0.93 |
| chr16_4069505_4069681 | 0.47 |
RP11-462G12.4 |
|
12420 |
0.19 |
| chr1_198904372_198904917 | 0.45 |
ENSG00000207759 |
. |
76362 |
0.11 |
| chr9_80262701_80263060 | 0.45 |
GNA14 |
guanine nucleotide binding protein (G protein), alpha 14 |
343 |
0.91 |
| chr12_29303115_29303266 | 0.45 |
FAR2 |
fatty acyl CoA reductase 2 |
1057 |
0.62 |
| chr5_5419342_5419575 | 0.44 |
KIAA0947 |
KIAA0947 |
3349 |
0.39 |
| chr4_1513980_1514175 | 0.44 |
NKX1-1 |
NK1 homeobox 1 |
113958 |
0.05 |
| chr2_98280710_98280922 | 0.43 |
LINC01125 |
long intergenic non-protein coding RNA 1125 |
81 |
0.64 |
| chr5_156570028_156570441 | 0.43 |
MED7 |
mediator complex subunit 7 |
39 |
0.7 |
| chr7_105494613_105494850 | 0.43 |
ATXN7L1 |
ataxin 7-like 1 |
22192 |
0.26 |
| chr10_65676733_65676884 | 0.43 |
REEP3 |
receptor accessory protein 3 |
395685 |
0.01 |
| chr14_93604450_93604601 | 0.42 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
21860 |
0.14 |
| chr20_57426029_57426307 | 0.42 |
GNAS-AS1 |
GNAS antisense RNA 1 |
210 |
0.89 |
| chr12_32832317_32832517 | 0.42 |
DNM1L |
dynamin 1-like |
147 |
0.97 |
| chr8_13370846_13370997 | 0.42 |
DLC1 |
deleted in liver cancer 1 |
1353 |
0.48 |
| chr12_94660827_94661045 | 0.41 |
PLXNC1 |
plexin C1 |
4639 |
0.19 |
| chr10_98443954_98444105 | 0.41 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
14661 |
0.21 |
| chr7_156432875_156433240 | 0.41 |
RNF32 |
ring finger protein 32 |
82 |
0.57 |
| chr20_53092813_53093232 | 0.40 |
DOK5 |
docking protein 5 |
765 |
0.8 |
| chr2_70140665_70140816 | 0.40 |
ENSG00000238465 |
. |
913 |
0.4 |
| chr17_63590380_63590998 | 0.39 |
AXIN2 |
axin 2 |
32924 |
0.2 |
| chr1_240816316_240816467 | 0.39 |
ENSG00000251750 |
. |
276 |
0.92 |
| chr4_159691379_159691530 | 0.39 |
FNIP2 |
folliculin interacting protein 2 |
1164 |
0.49 |
| chr22_42666087_42666289 | 0.39 |
TCF20 |
transcription factor 20 (AR1) |
54740 |
0.11 |
| chr11_47270476_47271047 | 0.38 |
NR1H3 |
nuclear receptor subfamily 1, group H, member 3 |
254 |
0.52 |
| chr2_9615136_9615532 | 0.38 |
IAH1 |
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae) |
150 |
0.95 |
| chr18_22864434_22864674 | 0.38 |
CTD-2006O16.2 |
|
17536 |
0.26 |
| chrX_48659526_48659734 | 0.38 |
HDAC6 |
histone deacetylase 6 |
154 |
0.91 |
| chr11_85429273_85429899 | 0.38 |
SYTL2 |
synaptotagmin-like 2 |
55 |
0.97 |
| chr12_131364699_131365309 | 0.37 |
RAN |
RAN, member RAS oncogene family |
8363 |
0.16 |
| chr8_90915010_90915840 | 0.37 |
OSGIN2 |
oxidative stress induced growth inhibitor family member 2 |
436 |
0.83 |
| chr22_28181066_28181439 | 0.37 |
MN1 |
meningioma (disrupted in balanced translocation) 1 |
16234 |
0.25 |
| chr12_68759315_68759468 | 0.37 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
33230 |
0.21 |
| chr2_231084808_231085018 | 0.37 |
SP110 |
SP110 nuclear body protein |
86 |
0.97 |
| chr16_51184162_51184313 | 0.36 |
SALL1 |
spalt-like transcription factor 1 |
271 |
0.87 |
| chr5_157893628_157893779 | 0.36 |
ENSG00000222626 |
. |
489739 |
0.0 |
| chr11_120468534_120468685 | 0.36 |
GRIK4 |
glutamate receptor, ionotropic, kainate 4 |
33466 |
0.21 |
| chr8_116681536_116682020 | 0.36 |
TRPS1 |
trichorhinophalangeal syndrome I |
68 |
0.99 |
| chrY_15591562_15591785 | 0.35 |
UTY |
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
122 |
0.98 |
| chr12_11323609_11324192 | 0.35 |
RP11-785H5.2 |
|
52 |
0.97 |
| chr2_107084278_107084429 | 0.35 |
RGPD3 |
RANBP2-like and GRIP domain containing 3 |
473 |
0.82 |
| chr7_5465601_5465997 | 0.35 |
TNRC18 |
trinucleotide repeat containing 18 |
754 |
0.57 |
| chr6_127835815_127835966 | 0.35 |
SOGA3 |
SOGA family member 3 |
1869 |
0.42 |
| chr9_35691198_35691509 | 0.35 |
TPM2 |
tropomyosin 2 (beta) |
336 |
0.72 |
| chr6_16586317_16586686 | 0.35 |
ENSG00000265642 |
. |
157747 |
0.04 |
| chr6_131382971_131383273 | 0.34 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
594 |
0.84 |
| chr15_26105615_26105919 | 0.34 |
ATP10A |
ATPase, class V, type 10A |
2588 |
0.28 |
| chr12_7848081_7848232 | 0.34 |
GDF3 |
growth differentiation factor 3 |
216 |
0.91 |
| chr17_40424420_40424618 | 0.34 |
AC003104.1 |
|
182 |
0.9 |
| chrX_128974400_128974551 | 0.34 |
ZDHHC9 |
zinc finger, DHHC-type containing 9 |
1325 |
0.44 |
| chr12_72665965_72666270 | 0.34 |
ENSG00000236333 |
. |
189 |
0.69 |
| chr14_81931293_81931471 | 0.34 |
STON2 |
stonin 2 |
28573 |
0.23 |
| chr19_1367548_1367699 | 0.34 |
AC004623.2 |
|
9614 |
0.08 |
| chr2_109151249_109151412 | 0.34 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
418 |
0.64 |
| chr2_102654423_102654574 | 0.34 |
IL1R1 |
interleukin 1 receptor, type I |
26506 |
0.19 |
| chr16_19728705_19729100 | 0.34 |
KNOP1 |
lysine-rich nucleolar protein 1 |
551 |
0.5 |
| chr13_24612503_24612731 | 0.34 |
SPATA13 |
spermatogenesis associated 13 |
58673 |
0.12 |
| chr2_108444784_108444988 | 0.34 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
1493 |
0.58 |
| chr4_79697241_79697410 | 0.33 |
RP11-109G23.3 |
|
198 |
0.5 |
| chr9_93570933_93571084 | 0.33 |
SYK |
spleen tyrosine kinase |
6799 |
0.34 |
| chr7_128809270_128809816 | 0.33 |
RP11-286H14.6 |
|
1490 |
0.31 |
| chr20_31260807_31260958 | 0.33 |
COMMD7 |
COMM domain containing 7 |
70330 |
0.08 |
| chr3_73769050_73769270 | 0.33 |
PDZRN3 |
PDZ domain containing ring finger 3 |
95069 |
0.08 |
| chr16_27412028_27412277 | 0.33 |
IL21R |
interleukin 21 receptor |
1331 |
0.47 |
| chr13_106704671_106704822 | 0.33 |
ENSG00000222643 |
. |
75974 |
0.12 |
| chr18_34273309_34273594 | 0.33 |
FHOD3 |
formin homology 2 domain containing 3 |
25069 |
0.22 |
| chr8_22342638_22342826 | 0.32 |
PPP3CC |
protein phosphatase 3, catalytic subunit, gamma isozyme |
9627 |
0.15 |
| chr14_90320865_90321016 | 0.32 |
EFCAB11 |
EF-hand calcium binding domain 11 |
99927 |
0.07 |
| chr6_114405713_114405864 | 0.32 |
RP3-399L15.3 |
|
55 |
0.98 |
| chr11_116658822_116659171 | 0.32 |
ZNF259 |
zinc finger protein 259 |
230 |
0.87 |
| chr5_88178054_88178448 | 0.32 |
MEF2C |
myocyte enhancer factor 2C |
713 |
0.52 |
| chr3_159706229_159707171 | 0.32 |
IL12A |
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
41 |
0.98 |
| chr14_92941981_92942132 | 0.32 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
36348 |
0.18 |
| chr12_85674997_85675148 | 0.32 |
ALX1 |
ALX homeobox 1 |
1187 |
0.65 |
| chr17_772598_772950 | 0.32 |
NXN |
nucleoredoxin |
5423 |
0.17 |
| chr5_149380800_149381041 | 0.32 |
TIGD6 |
tigger transposable element derived 6 |
190 |
0.82 |
| chr8_21770420_21771232 | 0.32 |
DOK2 |
docking protein 2, 56kDa |
348 |
0.88 |
| chr1_179335166_179335697 | 0.31 |
AXDND1 |
axonemal dynein light chain domain containing 1 |
324 |
0.87 |
| chr7_1578236_1578521 | 0.31 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
34 |
0.97 |
| chr20_58637452_58637603 | 0.31 |
C20orf197 |
chromosome 20 open reading frame 197 |
6547 |
0.27 |
| chr7_130645974_130646155 | 0.31 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
22804 |
0.23 |
| chr21_34863285_34863752 | 0.31 |
DNAJC28 |
DnaJ (Hsp40) homolog, subfamily C, member 28 |
262 |
0.87 |
| chr4_7832917_7833068 | 0.31 |
AFAP1 |
actin filament associated protein 1 |
40817 |
0.16 |
| chr15_77708252_77708450 | 0.31 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
4091 |
0.26 |
| chr1_43147265_43147870 | 0.31 |
YBX1 |
Y box binding protein 1 |
531 |
0.75 |
| chr4_56047702_56047853 | 0.31 |
ENSG00000239545 |
. |
34302 |
0.16 |
| chr1_202775672_202775981 | 0.31 |
KDM5B |
lysine (K)-specific demethylase 5B |
601 |
0.63 |
| chr12_123284077_123284228 | 0.31 |
CCDC62 |
coiled-coil domain containing 62 |
18365 |
0.13 |
| chr11_6411170_6411483 | 0.30 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
329 |
0.85 |
| chr4_95374257_95374632 | 0.30 |
PDLIM5 |
PDZ and LIM domain 5 |
1350 |
0.6 |
| chr11_72504918_72505117 | 0.30 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
291 |
0.55 |
| chrX_149905180_149905331 | 0.30 |
MTMR1 |
myotubularin related protein 1 |
18106 |
0.25 |
| chr7_87849519_87850007 | 0.30 |
SRI |
sorcin |
369 |
0.77 |
| chr19_53324249_53324569 | 0.30 |
ZNF28 |
zinc finger protein 28 |
476 |
0.77 |
| chr6_17212771_17212995 | 0.30 |
RBM24 |
RNA binding motif protein 24 |
68694 |
0.13 |
| chr4_71702728_71702883 | 0.29 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
1913 |
0.26 |
| chr2_114341836_114342054 | 0.29 |
WASH2P |
WAS protein family homolog 2 pseudogene |
281 |
0.85 |
| chr6_39759892_39760124 | 0.29 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
134 |
0.98 |
| chr2_85536945_85537100 | 0.29 |
ENSG00000221579 |
. |
10865 |
0.11 |
| chr6_117997172_117997416 | 0.29 |
NUS1 |
nuclear undecaprenyl pyrophosphate synthase 1 homolog (S. cerevisiae) |
629 |
0.81 |
| chr5_172296489_172296640 | 0.29 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
35150 |
0.14 |
| chrX_108975342_108975600 | 0.29 |
ACSL4 |
acyl-CoA synthetase long-chain family member 4 |
971 |
0.63 |
| chr2_26401607_26402234 | 0.29 |
GAREML |
GRB2 associated, regulator of MAPK1-like |
1665 |
0.38 |
| chr8_26370640_26370791 | 0.29 |
PNMA2 |
paraneoplastic Ma antigen 2 |
893 |
0.46 |
| chr4_154179778_154180018 | 0.29 |
TRIM2 |
tripartite motif containing 2 |
1336 |
0.41 |
| chr21_18983747_18983987 | 0.29 |
BTG3 |
BTG family, member 3 |
1295 |
0.49 |
| chr14_21540226_21540377 | 0.29 |
NDRG2 |
NDRG family member 2 |
1270 |
0.23 |
| chr20_34208992_34209143 | 0.29 |
SPAG4 |
sperm associated antigen 4 |
1560 |
0.25 |
| chr1_3563687_3563838 | 0.29 |
WRAP73 |
WD repeat containing, antisense to TP73 |
2828 |
0.16 |
| chr1_207226273_207226577 | 0.29 |
YOD1 |
YOD1 deubiquitinase |
100 |
0.61 |
| chr19_42745746_42746329 | 0.29 |
GSK3A |
glycogen synthase kinase 3 alpha |
8 |
0.92 |
| chr9_93970956_93971107 | 0.29 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
153146 |
0.04 |
| chr5_33892545_33892868 | 0.28 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
409 |
0.85 |
| chr17_7477286_7477712 | 0.28 |
EIF4A1 |
eukaryotic translation initiation factor 4A1 |
337 |
0.48 |
| chr7_136831101_136831290 | 0.28 |
hsa-mir-490 |
hsa-mir-490 |
17822 |
0.28 |
| chr10_31996328_31996772 | 0.28 |
ENSG00000222412 |
. |
48514 |
0.18 |
| chr8_123439435_123439791 | 0.28 |
ENSG00000238901 |
. |
243917 |
0.02 |
| chr9_14098246_14098397 | 0.28 |
NFIB |
nuclear factor I/B |
82470 |
0.11 |
| chr10_31446412_31446660 | 0.28 |
ENSG00000263578 |
. |
91560 |
0.08 |
| chr1_154150371_154150685 | 0.28 |
TPM3 |
tropomyosin 3 |
131 |
0.92 |
| chr10_13343824_13344114 | 0.28 |
PHYH |
phytanoyl-CoA 2-hydroxylase |
443 |
0.81 |
| chr11_123374923_123375196 | 0.28 |
GRAMD1B |
GRAM domain containing 1B |
21285 |
0.22 |
| chr19_15752519_15752670 | 0.28 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
475 |
0.8 |
| chr2_201577972_201578363 | 0.28 |
AOX1 |
aldehyde oxidase 1 |
50537 |
0.11 |
| chr11_35965857_35966431 | 0.28 |
LDLRAD3 |
low density lipoprotein receptor class A domain containing 3 |
516 |
0.78 |
| chr3_99369768_99369919 | 0.28 |
COL8A1 |
collagen, type VIII, alpha 1 |
12389 |
0.25 |
| chr5_180234522_180234938 | 0.28 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
1028 |
0.5 |
| chr11_65121571_65122031 | 0.28 |
TIGD3 |
tigger transposable element derived 3 |
437 |
0.71 |
| chr19_13044137_13044551 | 0.28 |
FARSA |
phenylalanyl-tRNA synthetase, alpha subunit |
150 |
0.86 |
| chr1_42801336_42801700 | 0.27 |
FOXJ3 |
forkhead box J3 |
30 |
0.97 |
| chr11_60611740_60611891 | 0.27 |
RP11-804A23.2 |
|
1377 |
0.22 |
| chr12_49525985_49526136 | 0.27 |
RP11-386G11.10 |
|
567 |
0.36 |
| chr13_101173468_101173697 | 0.27 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
5901 |
0.19 |
| chr4_134071254_134071890 | 0.27 |
PCDH10 |
protocadherin 10 |
1102 |
0.7 |
| chr7_5184079_5184242 | 0.27 |
ZNF890P |
zinc finger protein 890, pseudogene |
16734 |
0.17 |
| chr16_16092499_16092731 | 0.27 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
11098 |
0.24 |
| chr7_142490891_142491049 | 0.26 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
9839 |
0.18 |
| chr6_8101764_8102105 | 0.26 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
345 |
0.91 |
| chr3_46035020_46035171 | 0.26 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
2212 |
0.3 |
| chr6_36952765_36953398 | 0.26 |
MTCH1 |
mitochondrial carrier 1 |
752 |
0.63 |
| chr4_141419201_141419573 | 0.26 |
RP11-542P2.1 |
Glycosyltransferase 54 domain-containing protein |
11 |
0.98 |
| chr11_73019949_73020282 | 0.26 |
RP11-800A3.7 |
|
291 |
0.76 |
| chr12_62860096_62860418 | 0.26 |
MON2 |
MON2 homolog (S. cerevisiae) |
340 |
0.89 |
| chr1_150779976_150780304 | 0.26 |
CTSK |
cathepsin K |
230 |
0.9 |
| chrX_120337999_120338191 | 0.26 |
ENSG00000212032 |
. |
128397 |
0.05 |
| chr3_8359644_8359795 | 0.26 |
ENSG00000253081 |
. |
46615 |
0.19 |
| chr2_208482240_208482391 | 0.26 |
METTL21A |
methyltransferase like 21A |
6789 |
0.2 |
| chr8_10696654_10696859 | 0.26 |
PINX1 |
PIN2/TERF1 interacting, telomerase inhibitor 1 |
525 |
0.45 |
| chr2_232826006_232826949 | 0.26 |
DIS3L2 |
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
59 |
0.98 |
| chr12_21926599_21927146 | 0.26 |
KCNJ8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
883 |
0.57 |
| chr19_32006893_32007044 | 0.26 |
ENSG00000252780 |
. |
139399 |
0.05 |
| chr3_14513811_14513962 | 0.25 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
136 |
0.97 |
| chr22_41844915_41845188 | 0.25 |
TOB2 |
transducer of ERBB2, 2 |
2024 |
0.25 |
| chr19_3186026_3186436 | 0.25 |
NCLN |
nicalin |
297 |
0.85 |
| chr3_47324358_47324645 | 0.25 |
KLHL18 |
kelch-like family member 18 |
64 |
0.64 |
| chr6_35635788_35636125 | 0.25 |
ENSG00000265527 |
. |
3390 |
0.18 |
| chr5_131131799_131132359 | 0.25 |
FNIP1 |
folliculin interacting protein 1 |
535 |
0.5 |
| chr5_111510621_111510791 | 0.25 |
CTC-459M5.1 |
|
1439 |
0.4 |
| chr1_1293449_1293856 | 0.25 |
MXRA8 |
matrix-remodelling associated 8 |
263 |
0.78 |
| chr7_102157613_102157887 | 0.25 |
RASA4B |
RAS p21 protein activator 4B |
407 |
0.7 |
| chr20_55936781_55936932 | 0.25 |
MTRNR2L3 |
MT-RNR2-like 3 |
1978 |
0.23 |
| chr1_52083072_52083707 | 0.25 |
OSBPL9 |
oxysterol binding protein-like 9 |
608 |
0.77 |
| chr2_198120374_198120867 | 0.25 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
46623 |
0.12 |
| chr10_14879452_14879992 | 0.25 |
CDNF |
cerebral dopamine neurotrophic factor |
349 |
0.56 |
| chr2_175854784_175854957 | 0.25 |
ENSG00000201425 |
. |
12339 |
0.15 |
| chr9_35646178_35646825 | 0.25 |
RP11-331F9.4 |
|
234 |
0.81 |
| chr11_5711907_5712253 | 0.25 |
TRIM22 |
tripartite motif containing 22 |
177 |
0.91 |
| chr7_91510205_91510446 | 0.25 |
MTERF |
mitochondrial transcription termination factor |
291 |
0.94 |
| chr12_96529900_96530051 | 0.25 |
ENSG00000266889 |
. |
33948 |
0.15 |
| chr17_16472637_16472847 | 0.25 |
ZNF287 |
zinc finger protein 287 |
222 |
0.93 |
| chr11_2468791_2468942 | 0.25 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
2645 |
0.21 |
| chr18_47793998_47794618 | 0.25 |
CCDC11 |
coiled-coil domain containing 11 |
1416 |
0.36 |
| chr15_80445342_80445551 | 0.25 |
FAH |
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
207 |
0.95 |
| chr11_77530634_77531102 | 0.25 |
RSF1 |
remodeling and spacing factor 1 |
884 |
0.42 |
| chr5_68903659_68903865 | 0.24 |
RP11-848G14.2 |
|
10378 |
0.18 |
| chr16_87498544_87498873 | 0.24 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
26943 |
0.15 |
| chr17_1686722_1687045 | 0.24 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
11870 |
0.11 |
| chr14_50863724_50864050 | 0.24 |
CDKL1 |
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
235 |
0.94 |
| chr3_160281811_160282588 | 0.24 |
KPNA4 |
karyopherin alpha 4 (importin alpha 3) |
1177 |
0.52 |
| chr1_168329622_168329773 | 0.24 |
ENSG00000207974 |
. |
15065 |
0.23 |
| chr17_7464555_7465071 | 0.24 |
SENP3 |
SUMO1/sentrin/SMT3 specific peptidase 3 |
459 |
0.5 |
| chr20_48294953_48295215 | 0.24 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
35331 |
0.15 |
| chr7_112578777_112579037 | 0.24 |
C7orf60 |
chromosome 7 open reading frame 60 |
766 |
0.77 |
| chr6_149637823_149638060 | 0.24 |
TAB2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
1122 |
0.52 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.5 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 0.3 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.1 | 0.3 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.1 | 0.4 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.3 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.3 | GO:0010661 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 0.4 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.1 | 0.2 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.4 | GO:1903020 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.1 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.5 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.2 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.2 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.1 | 0.2 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.3 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.2 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.1 | 0.4 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.1 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.1 | 0.2 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.1 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.2 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.1 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.1 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.1 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.2 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0050942 | regulation of melanocyte differentiation(GO:0045634) positive regulation of melanocyte differentiation(GO:0045636) regulation of pigment cell differentiation(GO:0050932) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.2 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.4 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.2 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.2 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.8 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.2 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.2 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0021610 | cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0021561 | facial nerve development(GO:0021561) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.3 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.1 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.2 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0042416 | dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.1 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.4 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0031645 | negative regulation of neurological system process(GO:0031645) negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.3 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.2 | GO:0048194 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.0 | GO:0046321 | positive regulation of fatty acid oxidation(GO:0046321) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.0 | 0.1 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.0 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0090181 | regulation of cholesterol metabolic process(GO:0090181) |
| 0.0 | 0.0 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.4 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.2 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.2 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.2 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.0 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.2 | GO:0048489 | synaptic vesicle transport(GO:0048489) synaptic vesicle localization(GO:0097479) establishment of synaptic vesicle localization(GO:0097480) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.1 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) |
| 0.0 | 0.1 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.0 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) regulation of gastric acid secretion(GO:0060453) |
| 0.0 | 0.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.1 | GO:0007512 | adult heart development(GO:0007512) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:1901796 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.0 | 0.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.6 | GO:0010827 | regulation of glucose transport(GO:0010827) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.0 | GO:0051307 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.0 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0009414 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.0 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:0052509 | modulation by symbiont of host defense response(GO:0052031) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.0 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0033483 | gas homeostasis(GO:0033483) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.4 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0061202 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 1.0 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0043656 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0044438 | microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.0 | 0.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.0 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.5 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.4 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 0.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.2 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.1 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.1 | 0.2 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.3 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.1 | 0.3 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.1 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.2 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.1 | GO:0032357 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.6 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.5 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.6 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.3 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.8 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0002060 | nucleobase binding(GO:0002054) purine nucleobase binding(GO:0002060) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.0 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 1.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.0 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.2 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.5 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.6 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME INFLUENZA LIFE CYCLE | Genes involved in Influenza Life Cycle |
| 0.0 | 0.1 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.6 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.0 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.0 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME ADAPTIVE IMMUNE SYSTEM | Genes involved in Adaptive Immune System |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |