Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
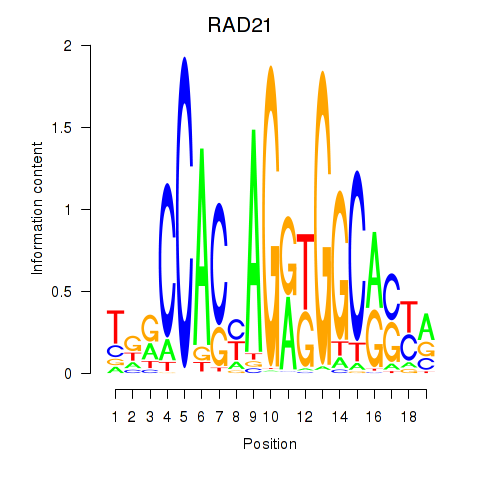
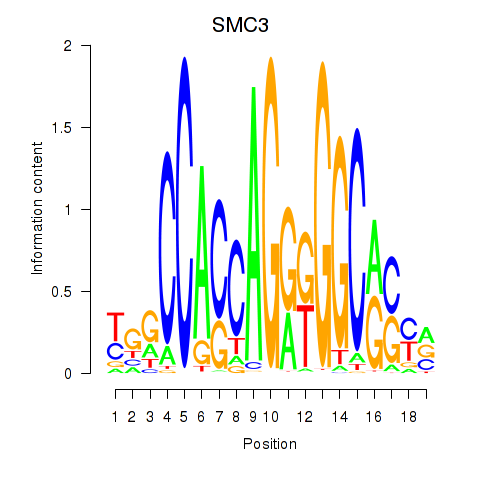
Results for RAD21_SMC3
Z-value: 1.50
Transcription factors associated with RAD21_SMC3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RAD21
|
ENSG00000164754.8 | RAD21 cohesin complex component |
|
SMC3
|
ENSG00000108055.9 | structural maintenance of chromosomes 3 |
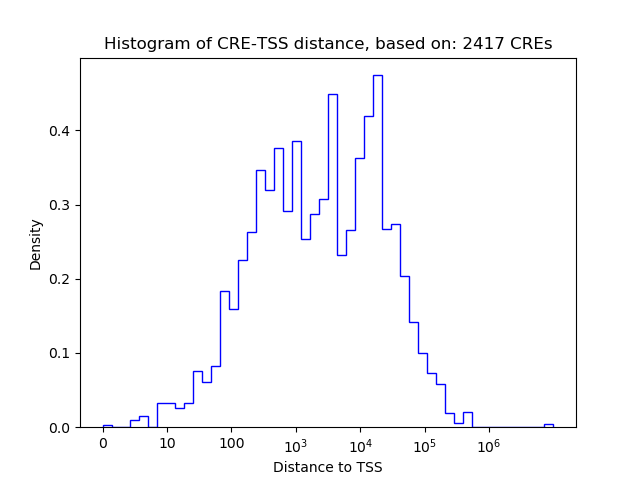
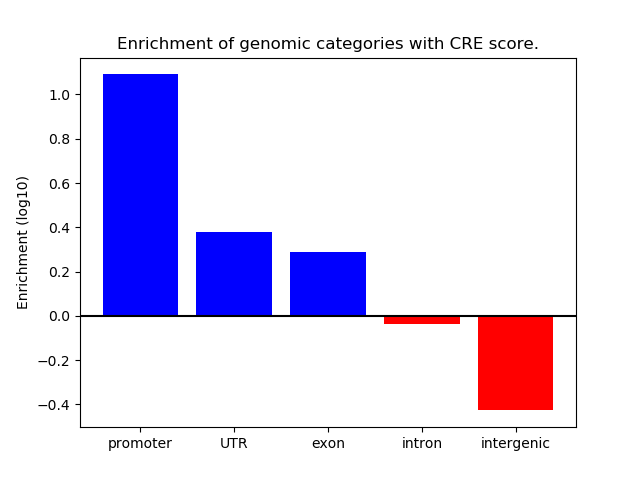
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_117887202_117887456 | RAD21 | 224 | 0.523990 | 0.62 | 7.5e-02 | Click! |
| chr8_117883179_117883330 | RAD21 | 3345 | 0.190759 | -0.25 | 5.1e-01 | Click! |
| chr8_117892142_117892293 | RAD21 | 5112 | 0.171511 | 0.24 | 5.3e-01 | Click! |
| chr8_117884616_117884915 | RAD21 | 1834 | 0.240314 | -0.21 | 5.8e-01 | Click! |
| chr8_117902849_117903000 | RAD21 | 15819 | 0.148365 | 0.19 | 6.2e-01 | Click! |
| chr10_112296305_112296649 | SMC3 | 30972 | 0.132365 | -0.56 | 1.1e-01 | Click! |
| chr10_112295759_112296296 | SMC3 | 31422 | 0.131407 | -0.47 | 2.1e-01 | Click! |
| chr10_112327463_112328125 | SMC3 | 345 | 0.864914 | 0.41 | 2.7e-01 | Click! |
| chr10_112327217_112327415 | SMC3 | 133 | 0.956732 | 0.28 | 4.7e-01 | Click! |
| chr10_112326670_112327053 | SMC3 | 588 | 0.721803 | 0.22 | 5.6e-01 | Click! |
Activity of the RAD21_SMC3 motif across conditions
Conditions sorted by the z-value of the RAD21_SMC3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
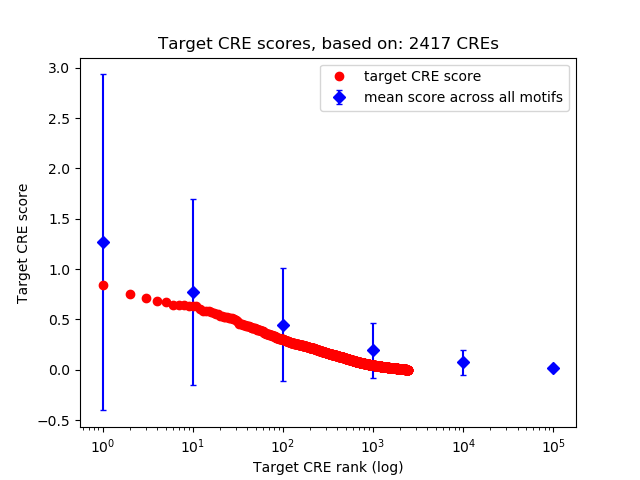
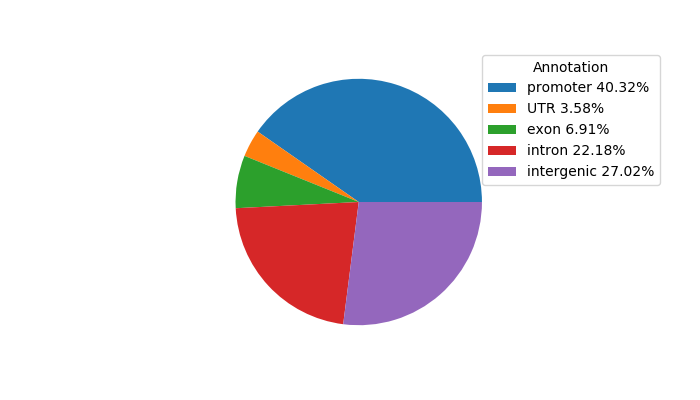
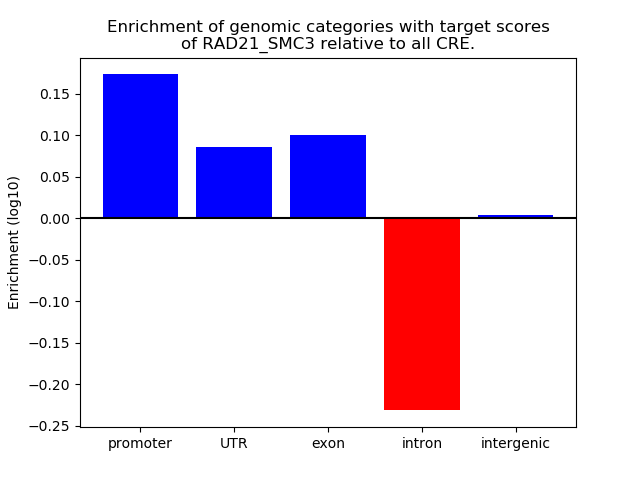
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_510720_510899 | 0.84 |
AC005775.2 |
|
2996 |
0.12 |
| chr14_103571364_103571515 | 0.75 |
EXOC3L4 |
exocyst complex component 3-like 4 |
2414 |
0.25 |
| chr19_4872699_4872850 | 0.71 |
ENSG00000264899 |
. |
3950 |
0.13 |
| chr16_23848456_23848777 | 0.69 |
PRKCB |
protein kinase C, beta |
72 |
0.98 |
| chr10_90595544_90595695 | 0.68 |
ANKRD22 |
ankyrin repeat domain 22 |
15956 |
0.16 |
| chr10_81903584_81903735 | 0.65 |
PLAC9 |
placenta-specific 9 |
11182 |
0.18 |
| chr19_7744309_7744460 | 0.64 |
CTD-3214H19.16 |
|
997 |
0.26 |
| chr11_728044_728195 | 0.64 |
AP006621.9 |
|
1072 |
0.28 |
| chr20_30410751_30410929 | 0.64 |
MYLK2 |
myosin light chain kinase 2 |
3664 |
0.17 |
| chr3_114582845_114583026 | 0.63 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
104817 |
0.08 |
| chr12_94543457_94543730 | 0.63 |
PLXNC1 |
plexin C1 |
1094 |
0.57 |
| chr17_63133333_63133484 | 0.60 |
RGS9 |
regulator of G-protein signaling 9 |
158 |
0.97 |
| chr1_33520793_33520944 | 0.59 |
AK2 |
adenylate kinase 2 |
18275 |
0.16 |
| chr19_41076676_41076827 | 0.58 |
SHKBP1 |
SH3KBP1 binding protein 1 |
6006 |
0.15 |
| chr1_152161161_152161312 | 0.58 |
FLG-AS1 |
FLG antisense RNA 1 |
591 |
0.7 |
| chr10_82295880_82296244 | 0.57 |
RP11-137H2.4 |
|
364 |
0.84 |
| chr19_42210710_42210868 | 0.56 |
CEACAM5 |
carcinoembryonic antigen-related cell adhesion molecule 5 |
1715 |
0.23 |
| chr5_39217983_39218134 | 0.56 |
FYB |
FYN binding protein |
1607 |
0.52 |
| chrX_40737289_40737555 | 0.56 |
MED14 |
mediator complex subunit 14 |
142639 |
0.04 |
| chr16_46663412_46663563 | 0.53 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
7949 |
0.18 |
| chr17_43212649_43212800 | 0.53 |
ACBD4 |
acyl-CoA binding domain containing 4 |
290 |
0.8 |
| chr17_63096707_63096868 | 0.53 |
RGS9 |
regulator of G-protein signaling 9 |
36779 |
0.14 |
| chr10_4891951_4892102 | 0.52 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
10050 |
0.23 |
| chr1_40265403_40265603 | 0.52 |
RP1-118J21.25 |
|
10855 |
0.13 |
| chr6_13278547_13278698 | 0.52 |
RP1-257A7.4 |
|
4331 |
0.22 |
| chr6_36259031_36259182 | 0.52 |
PNPLA1 |
patatin-like phospholipase domain containing 1 |
20869 |
0.16 |
| chr14_102990897_102991048 | 0.52 |
ANKRD9 |
ankyrin repeat domain 9 |
14836 |
0.15 |
| chr7_142937826_142938131 | 0.50 |
GSTK1 |
glutathione S-transferase kappa 1 |
3208 |
0.14 |
| chr22_37595607_37595758 | 0.50 |
SSTR3 |
somatostatin receptor 3 |
11323 |
0.12 |
| chr11_57226259_57226422 | 0.49 |
RTN4RL2 |
reticulon 4 receptor-like 2 |
1682 |
0.2 |
| chr19_47808671_47808977 | 0.48 |
C5AR1 |
complement component 5a receptor 1 |
4307 |
0.16 |
| chr1_26648089_26648240 | 0.48 |
UBXN11 |
UBX domain protein 11 |
3310 |
0.14 |
| chr12_54399885_54400449 | 0.46 |
HOXC8 |
homeobox C8 |
2665 |
0.09 |
| chr10_70853201_70853352 | 0.46 |
SRGN |
serglycin |
5402 |
0.21 |
| chr6_159463785_159463936 | 0.46 |
TAGAP |
T-cell activation RhoGTPase activating protein |
2190 |
0.31 |
| chr20_36919775_36919926 | 0.45 |
BPI |
bactericidal/permeability-increasing protein |
12675 |
0.15 |
| chr11_65149415_65150138 | 0.45 |
SLC25A45 |
solute carrier family 25, member 45 |
68 |
0.95 |
| chr12_131303541_131303831 | 0.44 |
ENSG00000238822 |
. |
3788 |
0.22 |
| chr20_31886970_31887121 | 0.44 |
BPIFB1 |
BPI fold containing family B, member 1 |
16104 |
0.16 |
| chr11_111802560_111802798 | 0.44 |
ENSG00000238965 |
. |
3555 |
0.13 |
| chr22_42062744_42062895 | 0.43 |
ENSG00000207457 |
. |
12896 |
0.1 |
| chr11_67381317_67381845 | 0.43 |
NDUFV1 |
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa |
5884 |
0.1 |
| chr5_118670778_118671030 | 0.43 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2034 |
0.35 |
| chr2_176944534_176944961 | 0.42 |
EVX2 |
even-skipped homeobox 2 |
3894 |
0.12 |
| chr22_24819614_24820020 | 0.42 |
ADORA2A |
adenosine A2a receptor |
218 |
0.94 |
| chr18_75362846_75362997 | 0.42 |
ENSG00000252260 |
. |
212602 |
0.02 |
| chr11_113906864_113907142 | 0.42 |
ZBTB16 |
zinc finger and BTB domain containing 16 |
23312 |
0.18 |
| chrX_48774424_48774575 | 0.42 |
PIM2 |
pim-2 oncogene |
1487 |
0.23 |
| chr19_11647018_11647169 | 0.41 |
CNN1 |
calponin 1, basic, smooth muscle |
2439 |
0.14 |
| chr8_124934794_124934968 | 0.41 |
FER1L6 |
fer-1-like 6 (C. elegans) |
33344 |
0.2 |
| chr3_16925560_16925711 | 0.40 |
PLCL2 |
phospholipase C-like 2 |
817 |
0.73 |
| chr6_136547532_136547683 | 0.40 |
RP13-143G15.4 |
|
874 |
0.62 |
| chr17_76133551_76133702 | 0.40 |
TMC6 |
transmembrane channel-like 6 |
5138 |
0.12 |
| chr14_104367537_104367688 | 0.40 |
C14orf2 |
chromosome 14 open reading frame 2 |
20219 |
0.13 |
| chrX_40403109_40403372 | 0.40 |
ENSG00000238730 |
. |
8425 |
0.22 |
| chr15_85114143_85114405 | 0.39 |
LINC00933 |
long intergenic non-protein coding RNA 933 |
119 |
0.76 |
| chr17_36762681_36762941 | 0.39 |
SRCIN1 |
SRC kinase signaling inhibitor 1 |
628 |
0.62 |
| chr8_57470509_57470758 | 0.39 |
RP11-17A4.2 |
|
68976 |
0.13 |
| chr7_27219071_27219572 | 0.39 |
RP1-170O19.20 |
Uncharacterized protein |
311 |
0.56 |
| chr22_22292451_22292602 | 0.38 |
LL22NC03-86G7.1 |
|
83 |
0.83 |
| chr1_150533351_150533580 | 0.38 |
ADAMTSL4-AS1 |
ADAMTSL4 antisense RNA 1 |
504 |
0.58 |
| chr13_46734728_46734988 | 0.37 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
7796 |
0.17 |
| chr2_114006851_114007002 | 0.36 |
ENSG00000189223 |
. |
1417 |
0.35 |
| chr22_24836933_24837084 | 0.36 |
ADORA2A |
adenosine A2a receptor |
8200 |
0.18 |
| chr19_2475398_2475867 | 0.36 |
GADD45B |
growth arrest and DNA-damage-inducible, beta |
493 |
0.74 |
| chr13_34116896_34117047 | 0.36 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
192204 |
0.03 |
| chr16_89307277_89307470 | 0.36 |
RP11-46C24.6 |
|
12273 |
0.11 |
| chr16_81771661_81771865 | 0.36 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
939 |
0.7 |
| chr10_95576079_95576230 | 0.35 |
LGI1 |
leucine-rich, glioma inactivated 1 |
58476 |
0.12 |
| chr4_185185188_185185495 | 0.35 |
ENSG00000221523 |
. |
3243 |
0.26 |
| chr4_185926512_185926756 | 0.35 |
HELT |
helt bHLH transcription factor |
13361 |
0.23 |
| chr11_62078778_62078929 | 0.35 |
SCGB1D4 |
secretoglobin, family 1D, member 4 |
12317 |
0.11 |
| chr2_96930696_96931115 | 0.34 |
TMEM127 |
transmembrane protein 127 |
827 |
0.4 |
| chr13_53542376_53542527 | 0.34 |
OLFM4 |
olfactomedin 4 |
60443 |
0.14 |
| chr21_42741228_42741619 | 0.34 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
589 |
0.77 |
| chr10_74454418_74454598 | 0.34 |
MCU |
mitochondrial calcium uniporter |
2131 |
0.29 |
| chr3_196411919_196412186 | 0.34 |
ENSG00000201441 |
. |
23783 |
0.11 |
| chrX_152200996_152201209 | 0.34 |
PNMA3 |
paraneoplastic Ma antigen 3 |
23664 |
0.12 |
| chr9_139923953_139924104 | 0.34 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
1288 |
0.16 |
| chr21_38362274_38362496 | 0.33 |
HLCS |
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
139 |
0.95 |
| chr19_39902575_39902778 | 0.33 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
549 |
0.52 |
| chr9_130547259_130547434 | 0.33 |
CDK9 |
cyclin-dependent kinase 9 |
612 |
0.4 |
| chr22_37656956_37657107 | 0.32 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
16543 |
0.14 |
| chr19_16293563_16293744 | 0.32 |
FAM32A |
family with sequence similarity 32, member A |
2538 |
0.18 |
| chr8_131314714_131314865 | 0.32 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
38394 |
0.21 |
| chr2_131090199_131090350 | 0.32 |
IMP4 |
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
9524 |
0.15 |
| chr22_32061724_32061875 | 0.32 |
PISD |
phosphatidylserine decarboxylase |
3381 |
0.25 |
| chr12_29360340_29360491 | 0.31 |
FAR2 |
fatty acyl CoA reductase 2 |
16183 |
0.25 |
| chr11_64637195_64637670 | 0.31 |
EHD1 |
EH-domain containing 1 |
5706 |
0.11 |
| chr2_231852070_231852221 | 0.31 |
SPATA3 |
spermatogenesis associated 3 |
8691 |
0.16 |
| chrX_118863929_118864080 | 0.31 |
SOWAHD |
sosondowah ankyrin repeat domain family member D |
28572 |
0.13 |
| chr6_134495232_134495950 | 0.31 |
SGK1 |
serum/glucocorticoid regulated kinase 1 |
411 |
0.86 |
| chr6_138725225_138725500 | 0.31 |
HEBP2 |
heme binding protein 2 |
6 |
0.98 |
| chrX_108956393_108956544 | 0.31 |
ACSL4 |
acyl-CoA synthetase long-chain family member 4 |
19974 |
0.21 |
| chr16_57452807_57452958 | 0.31 |
CCL17 |
chemokine (C-C motif) ligand 17 |
14203 |
0.12 |
| chr5_79603471_79603622 | 0.30 |
ENSG00000251828 |
. |
1359 |
0.41 |
| chr19_16493858_16494419 | 0.30 |
ENSG00000243745 |
. |
15632 |
0.14 |
| chr12_6647009_6647306 | 0.30 |
RP5-940J5.9 |
|
379 |
0.66 |
| chr19_14545137_14545294 | 0.30 |
PKN1 |
protein kinase N1 |
910 |
0.44 |
| chr20_56172175_56172326 | 0.30 |
ZBP1 |
Z-DNA binding protein 1 |
13071 |
0.21 |
| chr7_75622802_75623087 | 0.30 |
POR |
P450 (cytochrome) oxidoreductase |
11715 |
0.16 |
| chr2_68978993_68979334 | 0.30 |
ARHGAP25 |
Rho GTPase activating protein 25 |
17149 |
0.23 |
| chr1_154971510_154971950 | 0.30 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
3397 |
0.09 |
| chrX_43609757_43609908 | 0.30 |
MAOA |
monoamine oxidase A |
94284 |
0.09 |
| chr1_249101479_249101630 | 0.30 |
SH3BP5L |
SH3-binding domain protein 5-like |
9718 |
0.16 |
| chr1_47903418_47903665 | 0.29 |
FOXD2 |
forkhead box D2 |
1852 |
0.27 |
| chr12_94543061_94543212 | 0.29 |
PLXNC1 |
plexin C1 |
637 |
0.76 |
| chr3_52305451_52305846 | 0.29 |
ENSG00000199150 |
. |
3269 |
0.11 |
| chr19_46272674_46272825 | 0.29 |
SIX5 |
SIX homeobox 5 |
265 |
0.73 |
| chr16_66864896_66865106 | 0.29 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
101 |
0.95 |
| chr1_202968488_202968639 | 0.29 |
TMEM183A |
transmembrane protein 183A |
7951 |
0.12 |
| chr17_76418239_76418390 | 0.29 |
AC061992.1 |
Uncharacterized protein |
4095 |
0.17 |
| chr10_105127693_105127877 | 0.28 |
TAF5 |
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa |
61 |
0.96 |
| chr19_42302858_42303009 | 0.28 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
1848 |
0.24 |
| chr8_23315005_23315175 | 0.28 |
ENTPD4 |
ectonucleoside triphosphate diphosphohydrolase 4 |
70 |
0.97 |
| chr1_95320110_95320311 | 0.28 |
ENSG00000237416 |
. |
772 |
0.66 |
| chrX_52964079_52964300 | 0.28 |
FAM156A |
family with sequence similarity 156, member A |
21340 |
0.17 |
| chr14_75981526_75981933 | 0.27 |
BATF |
basic leucine zipper transcription factor, ATF-like |
7039 |
0.21 |
| chr11_8008537_8008688 | 0.27 |
EIF3F |
eukaryotic translation initiation factor 3, subunit F |
175 |
0.92 |
| chr6_159125269_159125420 | 0.27 |
SYTL3 |
synaptotagmin-like 3 |
41149 |
0.13 |
| chr19_49138973_49139430 | 0.27 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
1308 |
0.22 |
| chr1_212838734_212839243 | 0.27 |
ENSG00000207491 |
. |
26618 |
0.12 |
| chrX_54666271_54666506 | 0.27 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
109713 |
0.06 |
| chr2_24713646_24713947 | 0.27 |
NCOA1 |
nuclear receptor coactivator 1 |
1005 |
0.64 |
| chr9_81310465_81310616 | 0.27 |
PSAT1 |
phosphoserine aminotransferase 1 |
398481 |
0.01 |
| chr2_152204695_152204846 | 0.27 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
9336 |
0.17 |
| chr1_2126776_2126927 | 0.27 |
C1orf86 |
chromosome 1 open reading frame 86 |
638 |
0.58 |
| chr16_75032327_75032816 | 0.26 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
357 |
0.89 |
| chr19_41847643_41847794 | 0.26 |
TGFB1 |
transforming growth factor, beta 1 |
217 |
0.87 |
| chr3_49943854_49944005 | 0.26 |
CTD-2330K9.3 |
Uncharacterized protein |
2509 |
0.14 |
| chr11_48131630_48131789 | 0.26 |
ENSG00000263693 |
. |
13375 |
0.23 |
| chr11_20408753_20408968 | 0.26 |
PRMT3 |
protein arginine methyltransferase 3 |
216 |
0.95 |
| chr22_17652535_17653083 | 0.26 |
CECR5 |
cat eye syndrome chromosome region, candidate 5 |
6632 |
0.16 |
| chr12_124377548_124377699 | 0.26 |
DNAH10 |
dynein, axonemal, heavy chain 10 |
15237 |
0.11 |
| chr7_137583870_137584060 | 0.26 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
29050 |
0.18 |
| chr11_723305_723456 | 0.26 |
AP006621.9 |
|
3667 |
0.1 |
| chr14_50053552_50053772 | 0.26 |
RN7SL1 |
RNA, 7SL, cytoplasmic 1 |
364 |
0.55 |
| chr17_38474976_38475621 | 0.26 |
RARA |
retinoic acid receptor, alpha |
761 |
0.52 |
| chr14_105189941_105190399 | 0.26 |
ADSSL1 |
adenylosuccinate synthase like 1 |
353 |
0.81 |
| chr11_2172831_2172982 | 0.26 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
2073 |
0.18 |
| chr4_17550872_17551165 | 0.26 |
ENSG00000238536 |
. |
20458 |
0.13 |
| chr13_96130880_96131359 | 0.26 |
CLDN10 |
claudin 10 |
45261 |
0.16 |
| chr17_78997611_78997870 | 0.26 |
BAIAP2 |
BAI1-associated protein 2 |
11210 |
0.12 |
| chr9_19048749_19049283 | 0.26 |
RRAGA |
Ras-related GTP binding A |
356 |
0.81 |
| chr2_28825128_28825469 | 0.26 |
PLB1 |
phospholipase B1 |
512 |
0.83 |
| chr16_2390815_2390992 | 0.26 |
ABCA3 |
ATP-binding cassette, sub-family A (ABC1), member 3 |
156 |
0.92 |
| chrX_153978145_153979054 | 0.25 |
GAB3 |
GRB2-associated binding protein 3 |
733 |
0.55 |
| chr9_132244274_132244823 | 0.25 |
ENSG00000264298 |
. |
3713 |
0.27 |
| chr1_155942046_155942197 | 0.25 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
10 |
0.95 |
| chr10_90343073_90343515 | 0.25 |
RNLS |
renalase, FAD-dependent amine oxidase |
324 |
0.86 |
| chr22_17587383_17587676 | 0.25 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
14614 |
0.14 |
| chr1_149860084_149860235 | 0.25 |
HIST2H2AB |
histone cluster 2, H2ab |
693 |
0.28 |
| chr3_128564835_128565137 | 0.25 |
RP11-723O4.2 |
|
19461 |
0.16 |
| chr9_32572122_32572855 | 0.25 |
NDUFB6 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
669 |
0.62 |
| chr17_71817273_71817424 | 0.25 |
SDK2 |
sidekick cell adhesion molecule 2 |
177120 |
0.03 |
| chr1_9202388_9202539 | 0.25 |
ENSG00000207865 |
. |
9373 |
0.17 |
| chr14_103589198_103589361 | 0.25 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
519 |
0.76 |
| chr11_45864761_45864912 | 0.25 |
CRY2 |
cryptochrome 2 (photolyase-like) |
3833 |
0.19 |
| chr4_141174038_141174780 | 0.24 |
SCOC |
short coiled-coil protein |
4031 |
0.26 |
| chr10_134364622_134364773 | 0.24 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
13054 |
0.21 |
| chr12_124018329_124018683 | 0.24 |
RILPL1 |
Rab interacting lysosomal protein-like 1 |
241 |
0.89 |
| chr15_41198500_41199127 | 0.24 |
VPS18 |
vacuolar protein sorting 18 homolog (S. cerevisiae) |
12148 |
0.11 |
| chr16_67580972_67581123 | 0.24 |
FAM65A |
family with sequence similarity 65, member A |
2750 |
0.12 |
| chr5_43105962_43106287 | 0.24 |
ZNF131 |
zinc finger protein 131 |
14861 |
0.18 |
| chr1_145589458_145589609 | 0.24 |
NUDT17 |
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
94 |
0.94 |
| chr16_30441543_30441841 | 0.24 |
DCTPP1 |
dCTP pyrophosphatase 1 |
296 |
0.74 |
| chr6_26596047_26596198 | 0.24 |
ABT1 |
activator of basal transcription 1 |
1058 |
0.52 |
| chr8_41685722_41686425 | 0.24 |
ENSG00000241834 |
. |
11784 |
0.18 |
| chr11_104914891_104915295 | 0.24 |
CARD16 |
caspase recruitment domain family, member 16 |
941 |
0.54 |
| chr2_37502502_37502653 | 0.24 |
PRKD3 |
protein kinase D3 |
830 |
0.57 |
| chr1_221055703_221055854 | 0.24 |
HLX |
H2.0-like homeobox |
1194 |
0.46 |
| chr5_176882439_176883305 | 0.24 |
DBN1 |
drebrin 1 |
6518 |
0.1 |
| chr7_148663870_148664057 | 0.24 |
ENSG00000252316 |
. |
3556 |
0.17 |
| chr1_11779431_11780126 | 0.24 |
AGTRAP |
angiotensin II receptor-associated protein |
16363 |
0.11 |
| chr16_67193901_67194322 | 0.24 |
TRADD |
TNFRSF1A-associated via death domain |
90 |
0.63 |
| chr8_22551556_22552015 | 0.24 |
EGR3 |
early growth response 3 |
970 |
0.44 |
| chr17_74117442_74118068 | 0.24 |
EXOC7 |
exocyst complex component 7 |
98 |
0.95 |
| chr3_136732781_136732932 | 0.24 |
IL20RB-AS1 |
IL20RB antisense RNA 1 |
31818 |
0.19 |
| chr7_43878481_43878681 | 0.24 |
MRPS24 |
mitochondrial ribosomal protein S24 |
30575 |
0.14 |
| chr6_44225465_44225616 | 0.23 |
SLC35B2 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
249 |
0.84 |
| chr9_100955406_100955634 | 0.23 |
CORO2A |
coronin, actin binding protein, 2A |
598 |
0.77 |
| chr19_17416354_17416505 | 0.23 |
MRPL34 |
mitochondrial ribosomal protein L34 |
48 |
0.92 |
| chr12_52417641_52417792 | 0.23 |
NR4A1 |
nuclear receptor subfamily 4, group A, member 1 |
1100 |
0.39 |
| chr8_134307720_134307932 | 0.23 |
NDRG1 |
N-myc downstream regulated 1 |
953 |
0.67 |
| chr13_78052035_78052755 | 0.23 |
SCEL |
sciellin |
57414 |
0.14 |
| chr19_6481304_6482171 | 0.23 |
DENND1C |
DENN/MADD domain containing 1C |
27 |
0.95 |
| chr5_179517510_179517661 | 0.23 |
RNF130 |
ring finger protein 130 |
18467 |
0.19 |
| chr15_75077997_75078148 | 0.23 |
CSK |
c-src tyrosine kinase |
2813 |
0.16 |
| chr2_238330261_238330412 | 0.23 |
AC112721.1 |
Uncharacterized protein |
3583 |
0.24 |
| chr1_152432501_152432949 | 0.23 |
CRNN |
cornulin |
45986 |
0.09 |
| chr17_7146487_7146682 | 0.23 |
GABARAP |
GABA(A) receptor-associated protein |
495 |
0.51 |
| chr6_110026512_110026663 | 0.23 |
FIG4 |
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) |
3664 |
0.28 |
| chr2_25015571_25015910 | 0.22 |
CENPO |
centromere protein O |
274 |
0.69 |
| chr8_25041176_25041327 | 0.22 |
DOCK5 |
dedicator of cytokinesis 5 |
987 |
0.56 |
| chr19_43032396_43032749 | 0.22 |
CEACAM1 |
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
26 |
0.98 |
| chr17_7340388_7340539 | 0.22 |
RP11-104H15.9 |
|
379 |
0.55 |
| chr2_113384253_113384510 | 0.22 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
19053 |
0.16 |
| chr3_63964903_63965338 | 0.22 |
ATXN7 |
ataxin 7 |
11700 |
0.14 |
| chr1_21619720_21620176 | 0.22 |
RP5-1071N3.1 |
|
165 |
0.91 |
| chr7_155137689_155137840 | 0.22 |
BLACE |
B-cell acute lymphoblastic leukemia expressed |
22865 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.2 | 0.5 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.1 | 0.3 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.3 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 0.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.1 | 0.2 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.1 | 0.2 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.2 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.2 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.2 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.1 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.3 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.2 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.2 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0042635 | positive regulation of hair cycle(GO:0042635) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.2 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.2 | GO:0036473 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.2 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.3 | GO:0032770 | positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.2 | GO:1903078 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:1901984 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0060996 | dendritic spine development(GO:0060996) |
| 0.0 | 0.1 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0045408 | regulation of interleukin-6 biosynthetic process(GO:0045408) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.3 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0010642 | negative regulation of platelet-derived growth factor receptor signaling pathway(GO:0010642) |
| 0.0 | 0.1 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0031111 | negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.0 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:0032627 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.2 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.0 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.0 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0072162 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.0 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.0 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.2 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.2 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.0 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.3 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.1 | 0.2 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.1 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.2 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.2 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME GAB1 SIGNALOSOME | Genes involved in GAB1 signalosome |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |