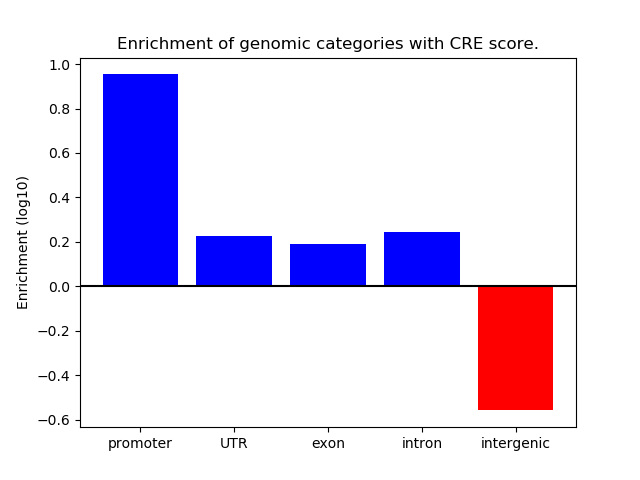
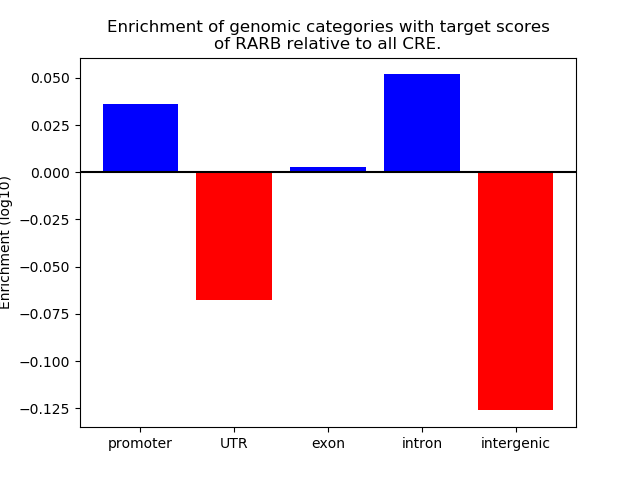
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
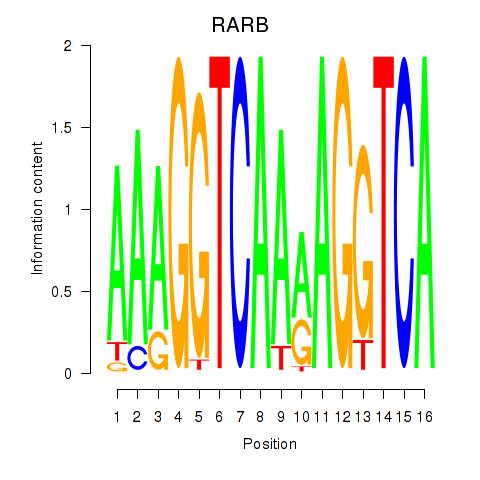
Results for RARB
Z-value: 1.32
Transcription factors associated with RARB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RARB
|
ENSG00000077092.14 | retinoic acid receptor beta |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_25496127_25496278 | RARB | 3599 | 0.335449 | -0.89 | 1.2e-03 | Click! |
| chr3_25497685_25497836 | RARB | 2041 | 0.437538 | -0.86 | 3.1e-03 | Click! |
| chr3_25469781_25469984 | RARB | 80 | 0.983222 | -0.86 | 3.2e-03 | Click! |
| chr3_25465504_25465718 | RARB | 4143 | 0.319253 | -0.86 | 3.3e-03 | Click! |
| chr3_25495752_25495903 | RARB | 3974 | 0.324844 | -0.85 | 3.9e-03 | Click! |
Activity of the RARB motif across conditions
Conditions sorted by the z-value of the RARB motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_98334070_98334346 | 0.86 |
ZAP70 |
zeta-chain (TCR) associated protein kinase 70kDa |
4185 |
0.2 |
| chr14_22961374_22961769 | 0.84 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
5400 |
0.11 |
| chr2_173327465_173327877 | 0.82 |
AC078883.3 |
|
3069 |
0.24 |
| chrX_70330282_70330478 | 0.81 |
IL2RG |
interleukin 2 receptor, gamma |
368 |
0.76 |
| chr6_143189309_143189460 | 0.77 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
31200 |
0.22 |
| chr6_152504718_152504928 | 0.73 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
15324 |
0.29 |
| chr3_183273082_183273407 | 0.73 |
KLHL6 |
kelch-like family member 6 |
233 |
0.92 |
| chr14_22950797_22950948 | 0.72 |
TRAJ55 |
T cell receptor alpha joining 55 (pseudogene) |
186 |
0.79 |
| chr17_76127167_76127361 | 0.71 |
TMC8 |
transmembrane channel-like 8 |
395 |
0.51 |
| chr13_52397765_52397916 | 0.68 |
RP11-327P2.5 |
|
19407 |
0.17 |
| chrX_11780001_11780185 | 0.66 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
2346 |
0.44 |
| chr6_128221812_128222507 | 0.66 |
THEMIS |
thymocyte selection associated |
56 |
0.99 |
| chr3_16329814_16329965 | 0.65 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
19141 |
0.15 |
| chr2_120687736_120687954 | 0.65 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
493 |
0.83 |
| chr14_22949623_22949774 | 0.64 |
ENSG00000251002 |
. |
580 |
0.49 |
| chr9_139990412_139990681 | 0.64 |
ENSG00000199411 |
. |
1749 |
0.12 |
| chr17_59981888_59982128 | 0.63 |
INTS2 |
integrator complex subunit 2 |
22779 |
0.17 |
| chr19_23984835_23985137 | 0.63 |
RP11-255H23.2 |
|
11412 |
0.19 |
| chr3_196368779_196369009 | 0.62 |
PIGX |
phosphatidylinositol glycan anchor biosynthesis, class X |
2248 |
0.2 |
| chr10_73843197_73843461 | 0.60 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
4757 |
0.26 |
| chrX_48772131_48772362 | 0.60 |
PIM2 |
pim-2 oncogene |
766 |
0.45 |
| chr22_37680309_37680627 | 0.59 |
CYTH4 |
cytohesin 4 |
1940 |
0.3 |
| chrX_78202204_78202379 | 0.58 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
1373 |
0.61 |
| chr2_175460517_175460758 | 0.58 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
1856 |
0.33 |
| chr3_121378552_121379115 | 0.58 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
912 |
0.5 |
| chr21_15917471_15917848 | 0.58 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
1003 |
0.63 |
| chr1_151129872_151130112 | 0.58 |
SCNM1 |
sodium channel modifier 1 |
852 |
0.29 |
| chr22_39639623_39639926 | 0.55 |
PDGFB |
platelet-derived growth factor beta polypeptide |
753 |
0.58 |
| chr13_99959249_99959863 | 0.55 |
GPR183 |
G protein-coupled receptor 183 |
103 |
0.97 |
| chr6_90788160_90788311 | 0.54 |
ENSG00000222078 |
. |
77010 |
0.1 |
| chr19_42061527_42061721 | 0.54 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
5738 |
0.18 |
| chrX_78406189_78406340 | 0.54 |
GPR174 |
G protein-coupled receptor 174 |
20205 |
0.29 |
| chr4_40202563_40202865 | 0.53 |
RHOH |
ras homolog family member H |
750 |
0.7 |
| chr5_130881605_130881983 | 0.53 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
13068 |
0.29 |
| chr7_138779169_138779413 | 0.53 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
14809 |
0.2 |
| chr13_75890123_75890274 | 0.53 |
TBC1D4 |
TBC1 domain family, member 4 |
25469 |
0.22 |
| chr8_8727283_8727434 | 0.52 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
23797 |
0.18 |
| chr10_14606149_14606418 | 0.52 |
FAM107B |
family with sequence similarity 107, member B |
7746 |
0.26 |
| chr2_87830138_87830289 | 0.52 |
RP11-1399P15.1 |
|
52660 |
0.16 |
| chr11_128387865_128388196 | 0.51 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
4066 |
0.27 |
| chr7_138778953_138779104 | 0.50 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
15015 |
0.2 |
| chr2_120988902_120989053 | 0.50 |
TMEM185B |
transmembrane protein 185B |
7993 |
0.17 |
| chr5_130849992_130850161 | 0.50 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
6226 |
0.32 |
| chr3_107844575_107844939 | 0.50 |
CD47 |
CD47 molecule |
34885 |
0.22 |
| chr18_56105832_56106078 | 0.49 |
ENSG00000207778 |
. |
12351 |
0.17 |
| chr2_112177530_112177803 | 0.49 |
ENSG00000266139 |
. |
98998 |
0.09 |
| chr19_10449910_10450285 | 0.48 |
ICAM3 |
intercellular adhesion molecule 3 |
198 |
0.86 |
| chr3_109525427_109525578 | 0.47 |
ENSG00000265956 |
. |
203827 |
0.03 |
| chr16_27414444_27414647 | 0.47 |
IL21R |
interleukin 21 receptor |
122 |
0.97 |
| chr2_213976639_213976790 | 0.47 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
36639 |
0.21 |
| chr5_156611761_156611912 | 0.47 |
ITK |
IL2-inducible T-cell kinase |
3999 |
0.16 |
| chr5_79484995_79485498 | 0.47 |
ENSG00000239159 |
. |
50162 |
0.12 |
| chr14_22996017_22996172 | 0.47 |
TRAJ15 |
T cell receptor alpha joining 15 |
2486 |
0.17 |
| chr1_32718292_32718517 | 0.46 |
LCK |
lymphocyte-specific protein tyrosine kinase |
1529 |
0.2 |
| chr12_123560353_123560523 | 0.46 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
204 |
0.94 |
| chr3_152000711_152000862 | 0.45 |
MBNL1-AS1 |
MBNL1 antisense RNA 1 |
13442 |
0.21 |
| chr6_15340960_15341111 | 0.45 |
ENSG00000201519 |
. |
16332 |
0.2 |
| chrX_153361307_153361458 | 0.44 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
1054 |
0.45 |
| chr12_22485870_22486021 | 0.44 |
ST8SIA1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
1152 |
0.64 |
| chr6_154567141_154567292 | 0.44 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
774 |
0.79 |
| chr15_26095881_26096079 | 0.43 |
ENSG00000266517 |
. |
2008 |
0.32 |
| chrX_78417451_78417673 | 0.43 |
GPR174 |
G protein-coupled receptor 174 |
8907 |
0.32 |
| chr9_138981896_138982047 | 0.43 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
5160 |
0.22 |
| chr20_62199083_62199345 | 0.43 |
HELZ2 |
helicase with zinc finger 2, transcriptional coactivator |
213 |
0.88 |
| chr13_74807358_74807666 | 0.43 |
ENSG00000206617 |
. |
55839 |
0.16 |
| chr17_10603321_10603720 | 0.43 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
2609 |
0.18 |
| chr1_154300198_154300512 | 0.42 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
79 |
0.94 |
| chr11_122722660_122722811 | 0.42 |
CRTAM |
cytotoxic and regulatory T cell molecule |
10276 |
0.21 |
| chr1_39684129_39684368 | 0.42 |
RP11-416A14.1 |
|
12064 |
0.17 |
| chr5_40515444_40515595 | 0.42 |
ENSG00000199552 |
. |
139546 |
0.05 |
| chr14_99735608_99735969 | 0.42 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
1777 |
0.38 |
| chr3_195267203_195267354 | 0.41 |
PPP1R2 |
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
2898 |
0.21 |
| chr1_198632029_198632471 | 0.41 |
RP11-553K8.5 |
|
3940 |
0.29 |
| chr11_128379892_128380047 | 0.41 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
4680 |
0.27 |
| chr2_225265820_225265971 | 0.41 |
FAM124B |
family with sequence similarity 124B |
816 |
0.76 |
| chr2_227703538_227703689 | 0.41 |
RHBDD1 |
rhomboid domain containing 1 |
1349 |
0.49 |
| chr2_231090336_231091061 | 0.40 |
SP140 |
SP140 nuclear body protein |
219 |
0.54 |
| chr2_201331503_201331739 | 0.40 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
26249 |
0.16 |
| chr8_21769513_21769765 | 0.40 |
DOK2 |
docking protein 2, 56kDa |
1535 |
0.4 |
| chr5_14676474_14676736 | 0.40 |
FAM105B |
family with sequence similarity 105, member B |
5054 |
0.24 |
| chr13_79967234_79967432 | 0.40 |
ENSG00000252496 |
. |
3567 |
0.23 |
| chr1_156785458_156785677 | 0.39 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
119 |
0.91 |
| chr7_123603687_123603838 | 0.39 |
SPAM1 |
sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) |
16008 |
0.19 |
| chr17_66196156_66196600 | 0.39 |
AMZ2 |
archaelysin family metallopeptidase 2 |
47337 |
0.11 |
| chr12_3861008_3861159 | 0.39 |
EFCAB4B |
EF-hand calcium binding domain 4B |
1180 |
0.56 |
| chr10_70444201_70444500 | 0.39 |
RP11-119F7.5 |
|
13907 |
0.15 |
| chr2_109237700_109238010 | 0.39 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
133 |
0.97 |
| chr3_128507459_128507964 | 0.39 |
ENSG00000244232 |
. |
3721 |
0.22 |
| chr16_87241773_87241924 | 0.39 |
RP11-899L11.3 |
|
7673 |
0.23 |
| chr6_11852612_11852763 | 0.39 |
ADTRP |
androgen-dependent TFPI-regulating protein |
45408 |
0.18 |
| chr17_4618141_4618477 | 0.39 |
ARRB2 |
arrestin, beta 2 |
575 |
0.53 |
| chr1_158978812_158979065 | 0.38 |
IFI16 |
interferon, gamma-inducible protein 16 |
153 |
0.96 |
| chr2_99386487_99386649 | 0.38 |
ENSG00000201070 |
. |
12305 |
0.21 |
| chr15_63483779_63483976 | 0.38 |
RAB8B |
RAB8B, member RAS oncogene family |
2072 |
0.34 |
| chr2_143886988_143887259 | 0.38 |
ARHGAP15 |
Rho GTPase activating protein 15 |
240 |
0.95 |
| chr7_87318553_87318704 | 0.38 |
ABCB1 |
ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
23936 |
0.21 |
| chr4_90224526_90224677 | 0.38 |
GPRIN3 |
GPRIN family member 3 |
4560 |
0.34 |
| chr15_61012992_61013143 | 0.38 |
ENSG00000212625 |
. |
15901 |
0.2 |
| chr12_15112961_15113206 | 0.38 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
1117 |
0.46 |
| chr2_212476372_212476523 | 0.38 |
ENSG00000199585 |
. |
76069 |
0.13 |
| chr15_81584764_81585010 | 0.37 |
IL16 |
interleukin 16 |
4367 |
0.24 |
| chr1_12533236_12533387 | 0.37 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
5293 |
0.24 |
| chr14_99710942_99711195 | 0.37 |
AL109767.1 |
|
18217 |
0.2 |
| chr7_104584232_104584453 | 0.37 |
ENSG00000251911 |
. |
27392 |
0.16 |
| chr21_46332173_46332515 | 0.37 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
969 |
0.4 |
| chr1_9882638_9884014 | 0.37 |
CLSTN1 |
calsyntenin 1 |
716 |
0.65 |
| chr19_50002038_50002189 | 0.36 |
ENSG00000200530 |
. |
1136 |
0.16 |
| chr19_50062782_50062933 | 0.36 |
NOSIP |
nitric oxide synthase interacting protein |
1030 |
0.27 |
| chr5_141467902_141468053 | 0.36 |
NDFIP1 |
Nedd4 family interacting protein 1 |
20093 |
0.2 |
| chr1_160547631_160547822 | 0.36 |
CD84 |
CD84 molecule |
1537 |
0.33 |
| chr21_30204306_30204701 | 0.36 |
N6AMT1 |
N-6 adenine-specific DNA methyltransferase 1 (putative) |
53190 |
0.14 |
| chr19_18414393_18414628 | 0.36 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
12559 |
0.09 |
| chr14_100534053_100534268 | 0.36 |
EVL |
Enah/Vasp-like |
1386 |
0.36 |
| chr1_12230241_12230523 | 0.36 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
3322 |
0.2 |
| chr3_71351252_71351403 | 0.35 |
FOXP1 |
forkhead box P1 |
2584 |
0.35 |
| chr1_100866151_100866481 | 0.35 |
ENSG00000216067 |
. |
21985 |
0.18 |
| chr9_36485594_36485745 | 0.35 |
RNF38 |
ring finger protein 38 |
84474 |
0.09 |
| chrX_40430227_40430433 | 0.35 |
ATP6AP2 |
ATPase, H+ transporting, lysosomal accessory protein 2 |
9816 |
0.21 |
| chr11_14722182_14722400 | 0.35 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
56914 |
0.14 |
| chr14_22947593_22947744 | 0.35 |
TRAJ60 |
T cell receptor alpha joining 60 (pseudogene) |
2372 |
0.15 |
| chr2_106392629_106392859 | 0.35 |
NCK2 |
NCK adaptor protein 2 |
30556 |
0.23 |
| chr17_56404642_56404793 | 0.35 |
BZRAP1 |
benzodiazepine receptor (peripheral) associated protein 1 |
728 |
0.47 |
| chr7_92744983_92745334 | 0.35 |
SAMD9 |
sterile alpha motif domain containing 9 |
2117 |
0.37 |
| chr1_207088744_207089040 | 0.35 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
6320 |
0.15 |
| chr6_128214596_128214747 | 0.35 |
THEMIS |
thymocyte selection associated |
7432 |
0.31 |
| chr9_117353596_117353986 | 0.34 |
ATP6V1G1 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G1 |
3765 |
0.18 |
| chr1_160635203_160635434 | 0.34 |
RP11-404F10.2 |
|
5250 |
0.17 |
| chr13_46753268_46753419 | 0.34 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
3116 |
0.21 |
| chr7_26704596_26704975 | 0.34 |
C7orf71 |
chromosome 7 open reading frame 71 |
27295 |
0.23 |
| chr22_44391651_44392034 | 0.34 |
PARVB |
parvin, beta |
3249 |
0.27 |
| chr19_16482178_16482409 | 0.34 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
9529 |
0.15 |
| chr1_24832184_24832335 | 0.34 |
RCAN3 |
RCAN family member 3 |
1798 |
0.32 |
| chr6_36171723_36172059 | 0.34 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
300 |
0.89 |
| chr14_32672059_32672251 | 0.33 |
ENSG00000202337 |
. |
320 |
0.8 |
| chr14_99737865_99738043 | 0.33 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
93 |
0.97 |
| chr6_154983150_154983301 | 0.33 |
SCAF8 |
SR-related CTD-associated factor 8 |
71234 |
0.12 |
| chr19_17136985_17137234 | 0.33 |
CPAMD8 |
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
516 |
0.7 |
| chr5_103287875_103288026 | 0.33 |
ENSG00000201910 |
. |
146625 |
0.05 |
| chr2_191883276_191883427 | 0.33 |
AC067945.2 |
|
671 |
0.6 |
| chrX_56792324_56792475 | 0.33 |
ENSG00000204272 |
. |
36707 |
0.23 |
| chr1_111162215_111162656 | 0.32 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
11661 |
0.19 |
| chr5_171610252_171610403 | 0.32 |
STK10 |
serine/threonine kinase 10 |
5063 |
0.21 |
| chr15_37350605_37350756 | 0.32 |
ENSG00000206676 |
. |
7549 |
0.24 |
| chr11_128566255_128566406 | 0.32 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
412 |
0.82 |
| chr13_100072035_100072186 | 0.32 |
ENSG00000266207 |
. |
33613 |
0.16 |
| chr11_128380151_128380303 | 0.32 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
4938 |
0.27 |
| chr1_29253197_29253478 | 0.32 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
12246 |
0.18 |
| chr2_8655557_8655708 | 0.32 |
AC011747.7 |
|
160264 |
0.04 |
| chr1_198632521_198632794 | 0.32 |
RP11-553K8.5 |
|
3533 |
0.3 |
| chr3_18484841_18485200 | 0.32 |
SATB1 |
SATB homeobox 1 |
1345 |
0.38 |
| chr2_68964970_68965234 | 0.31 |
ARHGAP25 |
Rho GTPase activating protein 25 |
3088 |
0.33 |
| chr11_118743547_118743698 | 0.31 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
10853 |
0.1 |
| chr7_129546585_129546878 | 0.31 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
44436 |
0.1 |
| chr1_160548800_160549253 | 0.31 |
CD84 |
CD84 molecule |
237 |
0.91 |
| chr10_26727488_26728306 | 0.31 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
543 |
0.85 |
| chr2_219762346_219762497 | 0.31 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
15538 |
0.1 |
| chr18_12838026_12838271 | 0.31 |
PTPN2 |
protein tyrosine phosphatase, non-receptor type 2 |
1325 |
0.51 |
| chr21_26945811_26946271 | 0.31 |
ENSG00000234883 |
. |
251 |
0.92 |
| chr18_60198500_60198651 | 0.30 |
ZCCHC2 |
zinc finger, CCHC domain containing 2 |
7723 |
0.28 |
| chr10_135203313_135203500 | 0.30 |
RP11-108K14.8 |
Mitochondrial GTPase 1 |
932 |
0.33 |
| chr5_79032666_79032916 | 0.30 |
CMYA5 |
cardiomyopathy associated 5 |
47091 |
0.16 |
| chr3_4867321_4867754 | 0.30 |
ENSG00000239126 |
. |
52849 |
0.14 |
| chr2_86080820_86081072 | 0.30 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
13831 |
0.17 |
| chr7_2559806_2560865 | 0.30 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
839 |
0.55 |
| chr20_5574610_5574761 | 0.30 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
16987 |
0.24 |
| chrX_147463091_147463259 | 0.30 |
AC002368.4 |
|
118960 |
0.06 |
| chr12_55376808_55376997 | 0.29 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
1280 |
0.52 |
| chr11_67008860_67009245 | 0.29 |
KDM2A |
lysine (K)-specific demethylase 2A |
1534 |
0.28 |
| chr20_47443528_47443935 | 0.29 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
689 |
0.79 |
| chr1_226921103_226921254 | 0.29 |
ITPKB |
inositol-trisphosphate 3-kinase B |
3981 |
0.28 |
| chr16_49497467_49497618 | 0.29 |
C16orf78 |
chromosome 16 open reading frame 78 |
89808 |
0.09 |
| chr4_71862773_71862924 | 0.29 |
DCK |
deoxycytidine kinase |
3493 |
0.32 |
| chr6_10529624_10529892 | 0.29 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
1169 |
0.49 |
| chr9_21444522_21444673 | 0.29 |
IFNA1 |
interferon, alpha 1 |
4157 |
0.18 |
| chr9_82239099_82239364 | 0.29 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
28277 |
0.28 |
| chr1_207090442_207090593 | 0.29 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
4695 |
0.16 |
| chr7_76541809_76541980 | 0.29 |
ENSG00000231183 |
. |
1472 |
0.46 |
| chr1_9690042_9690512 | 0.29 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
21513 |
0.15 |
| chr9_114836879_114837030 | 0.29 |
RP11-4O1.2 |
|
36944 |
0.14 |
| chr11_128161993_128162144 | 0.29 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
213221 |
0.02 |
| chr20_3137726_3137992 | 0.29 |
UBOX5 |
U-box domain containing 5 |
2663 |
0.16 |
| chr6_152506299_152506551 | 0.28 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
16926 |
0.28 |
| chr11_94801551_94801758 | 0.28 |
ENDOD1 |
endonuclease domain containing 1 |
21320 |
0.19 |
| chr2_10523080_10523233 | 0.28 |
HPCAL1 |
hippocalcin-like 1 |
36991 |
0.15 |
| chr8_22301822_22301996 | 0.28 |
PPP3CC |
protein phosphatase 3, catalytic subunit, gamma isozyme |
3031 |
0.21 |
| chr17_35860978_35861129 | 0.28 |
DUSP14 |
dual specificity phosphatase 14 |
8928 |
0.19 |
| chr17_80063977_80064793 | 0.28 |
CCDC57 |
coiled-coil domain containing 57 |
4659 |
0.1 |
| chr10_17726067_17726351 | 0.28 |
ENSG00000251959 |
. |
5022 |
0.18 |
| chr1_160174806_160174972 | 0.28 |
PEA15 |
phosphoprotein enriched in astrocytes 15 |
238 |
0.51 |
| chr4_109571201_109571501 | 0.28 |
OSTC |
oligosaccharyltransferase complex subunit (non-catalytic) |
389 |
0.83 |
| chr6_144980546_144980779 | 0.28 |
UTRN |
utrophin |
318 |
0.95 |
| chr6_45465042_45465193 | 0.28 |
RUNX2 |
runt-related transcription factor 2 |
74895 |
0.11 |
| chr15_32634653_32635232 | 0.28 |
ENSG00000221444 |
. |
50731 |
0.09 |
| chr10_5333412_5333586 | 0.28 |
AKR1C7P |
aldo-keto reductase family 1, member C7, pseudogene |
3066 |
0.27 |
| chr10_111817998_111818272 | 0.27 |
ADD3 |
adducin 3 (gamma) |
50413 |
0.13 |
| chr10_90519770_90519921 | 0.27 |
LIPN |
lipase, family member N |
1318 |
0.44 |
| chrX_70328757_70329094 | 0.27 |
IL2RG |
interleukin 2 receptor, gamma |
193 |
0.89 |
| chr16_67399178_67399329 | 0.27 |
LRRC36 |
leucine rich repeat containing 36 |
17947 |
0.09 |
| chr12_15104110_15104261 | 0.27 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
100 |
0.96 |
| chr9_77635535_77635794 | 0.27 |
RP11-197P3.5 |
|
4712 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.4 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.6 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.1 | 0.3 | GO:0072239 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.1 | 0.3 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.0 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.1 | 0.7 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.3 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.2 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.1 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.8 | GO:0045086 | positive regulation of interleukin-2 biosynthetic process(GO:0045086) |
| 0.1 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.1 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.2 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 0.2 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.1 | 0.2 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0002713 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.2 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.2 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0016242 | regulation of macroautophagy(GO:0016241) negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.5 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose transport(GO:0010829) negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.4 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0002834 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.4 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.2 | GO:0034383 | low-density lipoprotein particle clearance(GO:0034383) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.3 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.2 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:1902019 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:0070076 | histone lysine demethylation(GO:0070076) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0034391 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:1904181 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.1 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.0 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0002246 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0021894 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.7 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.0 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0001662 | behavioral fear response(GO:0001662) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.2 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.0 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.0 | GO:0042508 | tyrosine phosphorylation of Stat1 protein(GO:0042508) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.0 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.0 | GO:0032372 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.0 | 0.1 | GO:0051828 | entry into host cell(GO:0030260) entry into host(GO:0044409) entry into cell of other organism involved in symbiotic interaction(GO:0051806) entry into other organism involved in symbiotic interaction(GO:0051828) |
| 0.0 | 0.0 | GO:0043620 | regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0006386 | transcription elongation from RNA polymerase III promoter(GO:0006385) termination of RNA polymerase III transcription(GO:0006386) |
| 0.0 | 0.1 | GO:0032607 | interferon-alpha production(GO:0032607) regulation of interferon-alpha production(GO:0032647) positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 | 0.1 | GO:0032092 | positive regulation of protein binding(GO:0032092) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.1 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.0 | GO:0032715 | negative regulation of interleukin-6 production(GO:0032715) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.3 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0044217 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.2 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.1 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.2 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.2 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0032405 | MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.2 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.0 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.4 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.0 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 2.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.0 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.1 | 1.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.0 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |