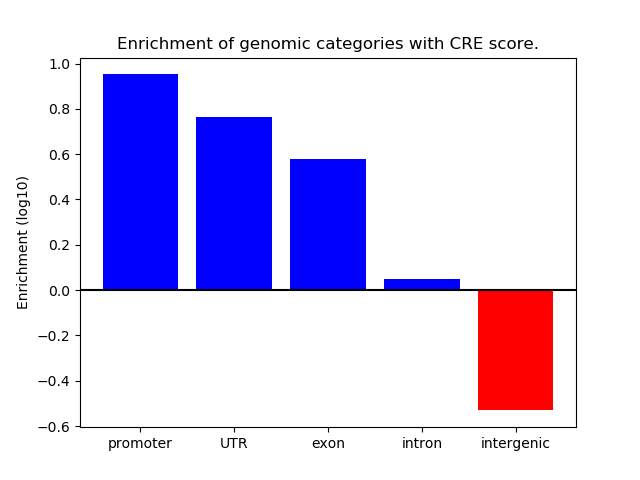
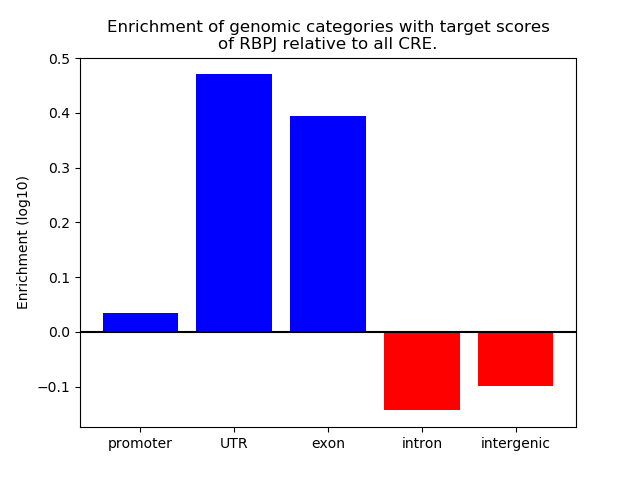
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
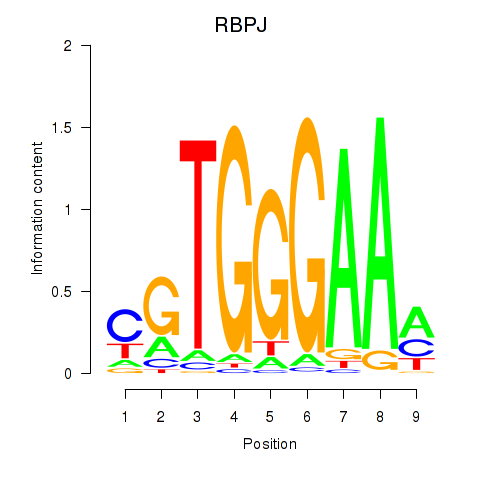
Results for RBPJ
Z-value: 0.45
Transcription factors associated with RBPJ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RBPJ
|
ENSG00000168214.16 | recombination signal binding protein for immunoglobulin kappa J region |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_26321348_26322561 | RBPJ | 231 | 0.964158 | -0.70 | 3.6e-02 | Click! |
| chr4_26208180_26208331 | RBPJ | 43178 | 0.209272 | -0.64 | 6.2e-02 | Click! |
| chr4_26095849_26096000 | RBPJ | 69153 | 0.133567 | 0.63 | 7.1e-02 | Click! |
| chr4_26301131_26301282 | RBPJ | 20078 | 0.283044 | -0.62 | 7.3e-02 | Click! |
| chr4_26325438_26325589 | RBPJ | 1021 | 0.699475 | -0.61 | 8.1e-02 | Click! |
Activity of the RBPJ motif across conditions
Conditions sorted by the z-value of the RBPJ motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
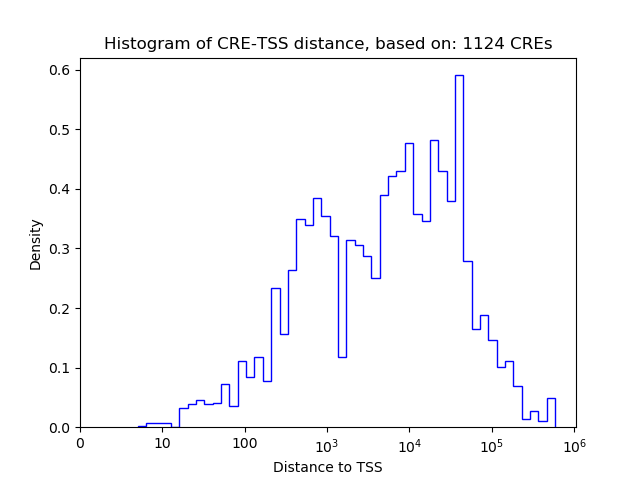
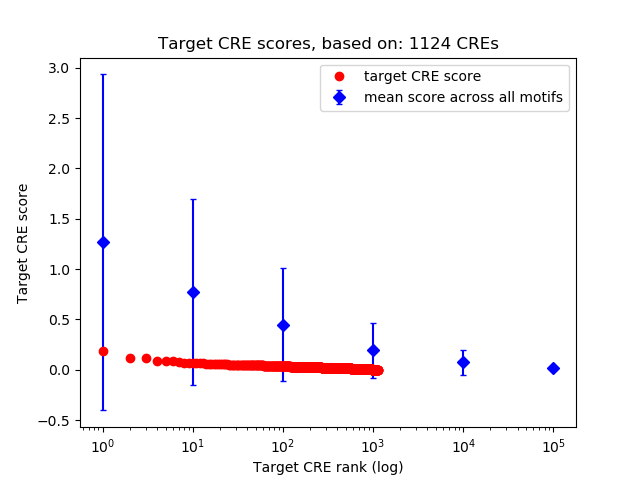
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_119953003_119953290 | 0.19 |
TRIM29 |
tripartite motif containing 29 |
38462 |
0.18 |
| chr11_119954900_119955051 | 0.12 |
TRIM29 |
tripartite motif containing 29 |
36633 |
0.19 |
| chr11_119954548_119954699 | 0.12 |
TRIM29 |
tripartite motif containing 29 |
36985 |
0.19 |
| chr5_63461313_63462499 | 0.09 |
RNF180 |
ring finger protein 180 |
83 |
0.99 |
| chr2_54682726_54683239 | 0.08 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
440 |
0.88 |
| chr7_99128907_99129569 | 0.08 |
FAM200A |
family with sequence similarity 200, member A |
20519 |
0.09 |
| chr6_27205685_27206711 | 0.07 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chrX_84499187_84500398 | 0.07 |
ZNF711 |
zinc finger protein 711 |
21 |
0.94 |
| chr19_10218994_10219145 | 0.06 |
ENSG00000209645 |
. |
742 |
0.32 |
| chr9_96108355_96109117 | 0.06 |
C9orf129 |
chromosome 9 open reading frame 129 |
40 |
0.98 |
| chr18_7116438_7116788 | 0.06 |
LAMA1 |
laminin, alpha 1 |
1200 |
0.55 |
| chr21_46875019_46875466 | 0.06 |
COL18A1 |
collagen, type XVIII, alpha 1 |
161 |
0.95 |
| chr19_9524094_9524245 | 0.06 |
ZNF266 |
zinc finger protein 266 |
5588 |
0.18 |
| chr13_78493097_78493683 | 0.06 |
EDNRB |
endothelin receptor type B |
424 |
0.48 |
| chr4_17618154_17618319 | 0.06 |
MED28 |
mediator complex subunit 28 |
1982 |
0.25 |
| chr3_64672065_64672629 | 0.06 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
1008 |
0.5 |
| chrX_130036036_130037401 | 0.06 |
ENOX2 |
ecto-NOX disulfide-thiol exchanger 2 |
460 |
0.9 |
| chr19_12502733_12502884 | 0.06 |
CTD-3105H18.4 |
|
8307 |
0.12 |
| chr3_140769768_140770253 | 0.06 |
SPSB4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
234 |
0.95 |
| chr4_176922498_176923243 | 0.06 |
GPM6A |
glycoprotein M6A |
613 |
0.78 |
| chr3_71586010_71586190 | 0.06 |
ENSG00000221264 |
. |
5140 |
0.23 |
| chr21_46530236_46530387 | 0.06 |
PRED58 |
|
4562 |
0.17 |
| chr2_105275388_105275738 | 0.05 |
ENSG00000207249 |
. |
74129 |
0.1 |
| chr21_35897414_35897754 | 0.05 |
RCAN1 |
regulator of calcineurin 1 |
102 |
0.97 |
| chr6_41605286_41606011 | 0.05 |
MDFI |
MyoD family inhibitor |
528 |
0.74 |
| chr8_145689855_145691101 | 0.05 |
CYHR1 |
cysteine/histidine-rich 1 |
60 |
0.88 |
| chr1_234041439_234041955 | 0.05 |
SLC35F3 |
solute carrier family 35, member F3 |
1018 |
0.65 |
| chr12_122460681_122461523 | 0.05 |
BCL7A |
B-cell CLL/lymphoma 7A |
1310 |
0.49 |
| chr2_159825200_159826543 | 0.05 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
688 |
0.73 |
| chr2_177392551_177392702 | 0.05 |
ENSG00000252027 |
. |
136778 |
0.04 |
| chr1_173175995_173176483 | 0.05 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
213 |
0.96 |
| chr4_30718989_30719310 | 0.05 |
PCDH7 |
protocadherin 7 |
2888 |
0.4 |
| chr8_146156057_146156445 | 0.05 |
ZNF16 |
zinc finger protein 16 |
19730 |
0.14 |
| chr4_118904778_118905073 | 0.05 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
50575 |
0.19 |
| chr19_11891380_11891531 | 0.05 |
ZNF441 |
zinc finger protein 441 |
13548 |
0.11 |
| chr9_8718682_8718836 | 0.05 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
15135 |
0.22 |
| chr16_31438814_31439148 | 0.05 |
COX6A2 |
cytochrome c oxidase subunit VIa polypeptide 2 |
768 |
0.46 |
| chr14_104582609_104583337 | 0.05 |
ENSG00000207568 |
. |
769 |
0.59 |
| chr6_24779816_24779967 | 0.05 |
GMNN |
geminin, DNA replication inhibitor |
2797 |
0.21 |
| chr6_72129322_72130506 | 0.05 |
ENSG00000207827 |
. |
16590 |
0.22 |
| chr16_3273990_3274238 | 0.05 |
ZNF200 |
zinc finger protein 200 |
10956 |
0.1 |
| chr4_57973648_57973950 | 0.05 |
ENSG00000238541 |
. |
1288 |
0.36 |
| chr18_25754690_25754841 | 0.05 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
2645 |
0.44 |
| chr19_58132756_58132907 | 0.05 |
ZNF134 |
zinc finger protein 134 |
7207 |
0.1 |
| chrX_47272886_47273037 | 0.05 |
ENSG00000221459 |
. |
24913 |
0.14 |
| chr13_24607852_24608071 | 0.05 |
SPATA13 |
spermatogenesis associated 13 |
54017 |
0.13 |
| chr19_12637235_12637523 | 0.05 |
CTD-2192J16.21 |
|
6408 |
0.12 |
| chr20_2260809_2260960 | 0.05 |
TGM3 |
transglutaminase 3 |
15763 |
0.23 |
| chr3_133710186_133710337 | 0.05 |
SLCO2A1 |
solute carrier organic anion transporter family, member 2A1 |
38497 |
0.17 |
| chr22_20119477_20120671 | 0.05 |
ZDHHC8 |
zinc finger, DHHC-type containing 8 |
603 |
0.53 |
| chr9_128652555_128652706 | 0.05 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
25080 |
0.27 |
| chr2_182675706_182676117 | 0.04 |
ENSG00000221023 |
. |
72578 |
0.09 |
| chr1_223351484_223351635 | 0.04 |
TLR5 |
toll-like receptor 5 |
34935 |
0.22 |
| chr16_69140450_69141374 | 0.04 |
HAS3 |
hyaluronan synthase 3 |
400 |
0.81 |
| chr17_5086107_5086258 | 0.04 |
ZNF594 |
zinc finger protein 594 |
1510 |
0.3 |
| chr18_908458_909079 | 0.04 |
RP11-672L10.3 |
|
1087 |
0.42 |
| chr2_28565345_28565858 | 0.04 |
AC093690.1 |
|
32275 |
0.15 |
| chr12_122884907_122885085 | 0.04 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
435 |
0.86 |
| chr4_11652300_11652527 | 0.04 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
221024 |
0.02 |
| chr19_58050211_58050362 | 0.04 |
ZNF550 |
zinc finger protein 550 |
9075 |
0.1 |
| chr19_12256353_12256531 | 0.04 |
ZNF625 |
zinc finger protein 625 |
2175 |
0.18 |
| chr4_187648163_187648374 | 0.04 |
FAT1 |
FAT atypical cadherin 1 |
392 |
0.92 |
| chr6_79902044_79902327 | 0.04 |
HMGN3-AS1 |
HMGN3 antisense RNA 1 |
41250 |
0.16 |
| chr4_186130212_186131033 | 0.04 |
KIAA1430 |
KIAA1430 |
36 |
0.92 |
| chr18_21198891_21199952 | 0.04 |
ANKRD29 |
ankyrin repeat domain 29 |
30065 |
0.14 |
| chr10_25836306_25836457 | 0.04 |
ENSG00000222543 |
. |
153608 |
0.04 |
| chr10_115860454_115861476 | 0.04 |
ENSG00000253066 |
. |
10208 |
0.2 |
| chr12_133780930_133781187 | 0.04 |
AC226150.4 |
Uncharacterized protein |
6714 |
0.13 |
| chr11_66153976_66154134 | 0.04 |
ENSG00000202317 |
. |
6865 |
0.08 |
| chr11_119953470_119953621 | 0.04 |
TRIM29 |
tripartite motif containing 29 |
38063 |
0.18 |
| chr11_8615266_8616201 | 0.04 |
STK33 |
serine/threonine kinase 33 |
26 |
0.97 |
| chr2_105475725_105475876 | 0.04 |
POU3F3 |
POU class 3 homeobox 3 |
3831 |
0.16 |
| chr10_72217935_72218559 | 0.04 |
NODAL |
nodal growth differentiation factor |
10540 |
0.17 |
| chr13_88329839_88330831 | 0.04 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
4837 |
0.33 |
| chr3_46263422_46263573 | 0.04 |
CCR1 |
chemokine (C-C motif) receptor 1 |
13610 |
0.2 |
| chr19_58774047_58774198 | 0.04 |
CTD-3138B18.6 |
|
3879 |
0.12 |
| chr19_46572119_46572325 | 0.04 |
IGFL4 |
IGF-like family member 4 |
8130 |
0.15 |
| chr19_58265433_58265584 | 0.04 |
ZNF776 |
zinc finger protein 776 |
3336 |
0.12 |
| chrX_19140154_19140583 | 0.04 |
GPR64 |
G protein-coupled receptor 64 |
211 |
0.97 |
| chr5_180277181_180277332 | 0.04 |
ZFP62 |
ZFP62 zinc finger protein |
10407 |
0.16 |
| chr19_460012_460518 | 0.04 |
SHC2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
731 |
0.5 |
| chr5_68788670_68789791 | 0.04 |
OCLN |
occludin |
612 |
0.62 |
| chr13_20175736_20176974 | 0.04 |
MPHOSPH8 |
M-phase phosphoprotein 8 |
31433 |
0.18 |
| chr1_218672576_218672996 | 0.04 |
C1orf143 |
chromosome 1 open reading frame 143 |
10652 |
0.26 |
| chr19_52469059_52469210 | 0.04 |
ZNF350 |
zinc finger protein 350 |
8972 |
0.12 |
| chr19_58945059_58945210 | 0.04 |
ZNF132 |
zinc finger protein 132 |
6455 |
0.08 |
| chr12_120142120_120142271 | 0.04 |
RP1-127H14.3 |
Uncharacterized protein |
4622 |
0.19 |
| chr8_61193398_61193986 | 0.04 |
CA8 |
carbonic anhydrase VIII |
279 |
0.95 |
| chr19_12016806_12016962 | 0.04 |
ZNF69 |
zinc finger protein 69 |
18214 |
0.1 |
| chr2_145210039_145210510 | 0.04 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
22137 |
0.25 |
| chr19_11833281_11833438 | 0.04 |
CTC-499B15.6 |
|
8239 |
0.13 |
| chr12_32135997_32136148 | 0.04 |
KIAA1551 |
KIAA1551 |
20649 |
0.22 |
| chr18_64307786_64307937 | 0.04 |
ENSG00000221536 |
. |
12877 |
0.26 |
| chr1_178063691_178064117 | 0.04 |
RASAL2 |
RAS protein activator like 2 |
628 |
0.84 |
| chr21_48087065_48088237 | 0.04 |
PRMT2 |
protein arginine methyltransferase 2 |
23396 |
0.22 |
| chr19_58773538_58773689 | 0.04 |
CTD-3138B18.6 |
|
4388 |
0.11 |
| chr7_5634981_5635198 | 0.04 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
772 |
0.6 |
| chr21_16424202_16424355 | 0.04 |
NRIP1 |
nuclear receptor interacting protein 1 |
12848 |
0.26 |
| chr7_93519153_93519436 | 0.04 |
TFPI2 |
tissue factor pathway inhibitor 2 |
188 |
0.88 |
| chr5_123985243_123985413 | 0.04 |
RP11-436H11.2 |
|
79196 |
0.08 |
| chr2_153325586_153326000 | 0.04 |
FMNL2 |
formin-like 2 |
134042 |
0.05 |
| chr9_99521835_99521986 | 0.04 |
ZNF510 |
zinc finger protein 510 |
18418 |
0.24 |
| chr6_72206174_72206487 | 0.03 |
ENSG00000212099 |
. |
42349 |
0.17 |
| chr2_232479090_232479920 | 0.03 |
C2orf57 |
chromosome 2 open reading frame 57 |
21930 |
0.15 |
| chr12_133779073_133779476 | 0.03 |
AC226150.4 |
Uncharacterized protein |
8498 |
0.13 |
| chr19_58689540_58689691 | 0.03 |
ZNF274 |
zinc finger protein 274 |
4781 |
0.15 |
| chr10_60273912_60274168 | 0.03 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
1140 |
0.65 |
| chr12_102225232_102225392 | 0.03 |
GNPTAB |
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
596 |
0.63 |
| chr2_103235205_103235939 | 0.03 |
SLC9A2 |
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
594 |
0.82 |
| chr2_121103939_121104858 | 0.03 |
INHBB |
inhibin, beta B |
679 |
0.78 |
| chr13_45146605_45146867 | 0.03 |
TSC22D1 |
TSC22 domain family, member 1 |
3656 |
0.33 |
| chr19_2933699_2933850 | 0.03 |
AC006277.2 |
|
6969 |
0.12 |
| chr10_18655851_18656002 | 0.03 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
26260 |
0.24 |
| chr12_13492233_13492384 | 0.03 |
C12orf36 |
chromosome 12 open reading frame 36 |
37056 |
0.18 |
| chr16_74734045_74735115 | 0.03 |
MLKL |
mixed lineage kinase domain-like |
108 |
0.97 |
| chr19_11892320_11892471 | 0.03 |
ZNF441 |
zinc finger protein 441 |
14488 |
0.11 |
| chr16_72459537_72459688 | 0.03 |
ENSG00000207514 |
. |
49009 |
0.17 |
| chr2_48476024_48476175 | 0.03 |
ENSG00000201010 |
. |
31385 |
0.2 |
| chr13_107572015_107572438 | 0.03 |
ENSG00000239050 |
. |
55148 |
0.17 |
| chr15_83679247_83679398 | 0.03 |
C15orf40 |
chromosome 15 open reading frame 40 |
1015 |
0.36 |
| chr1_224686385_224686769 | 0.03 |
WDR26 |
WD repeat domain 26 |
61842 |
0.11 |
| chr3_141131899_141132175 | 0.03 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
10377 |
0.23 |
| chr1_81137188_81137522 | 0.03 |
ENSG00000266033 |
. |
342182 |
0.01 |
| chr19_9639561_9639712 | 0.03 |
ZNF426 |
zinc finger protein 426 |
7609 |
0.17 |
| chr16_3432968_3433133 | 0.03 |
MTRNR2L4 |
MT-RNR2-like 4 |
10767 |
0.11 |
| chr5_4082877_4083178 | 0.03 |
CTD-2012M11.3 |
|
482725 |
0.01 |
| chr19_44514850_44515001 | 0.03 |
ZNF230 |
zinc finger protein 230 |
4073 |
0.12 |
| chr5_149524115_149524266 | 0.03 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
7190 |
0.15 |
| chr3_158508847_158509075 | 0.03 |
RP11-379F4.8 |
|
8625 |
0.16 |
| chr5_180278006_180278157 | 0.03 |
ZFP62 |
ZFP62 zinc finger protein |
9582 |
0.17 |
| chr3_119030384_119030661 | 0.03 |
ARHGAP31-AS1 |
ARHGAP31 antisense RNA 1 |
11085 |
0.16 |
| chr9_110013679_110013830 | 0.03 |
RAD23B |
RAD23 homolog B (S. cerevisiae) |
31664 |
0.23 |
| chr17_5085116_5085307 | 0.03 |
ZNF594 |
zinc finger protein 594 |
2481 |
0.2 |
| chr5_159797706_159797884 | 0.03 |
C1QTNF2 |
C1q and tumor necrosis factor related protein 2 |
147 |
0.95 |
| chr6_10586335_10586486 | 0.03 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
431 |
0.84 |
| chr10_98455004_98455155 | 0.03 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
25192 |
0.18 |
| chr5_58958074_58958460 | 0.03 |
ENSG00000202601 |
. |
41262 |
0.2 |
| chr15_75339241_75339413 | 0.03 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
3711 |
0.2 |
| chr11_12063788_12063939 | 0.03 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
32547 |
0.2 |
| chr7_41740779_41741576 | 0.03 |
INHBA |
inhibin, beta A |
970 |
0.56 |
| chr1_236046770_236047092 | 0.03 |
LYST |
lysosomal trafficking regulator |
59 |
0.97 |
| chr5_124079913_124080218 | 0.03 |
ZNF608 |
zinc finger protein 608 |
149 |
0.94 |
| chr17_47987589_47988019 | 0.03 |
ENSG00000199492 |
. |
55736 |
0.08 |
| chr17_60885608_60885921 | 0.03 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
59 |
0.98 |
| chr10_91274995_91275346 | 0.03 |
SLC16A12 |
solute carrier family 16, member 12 |
20143 |
0.18 |
| chr16_3108546_3109349 | 0.03 |
RP11-473M20.7 |
|
139 |
0.86 |
| chr1_21900562_21900713 | 0.03 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
334 |
0.87 |
| chr5_9544908_9545592 | 0.03 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
937 |
0.56 |
| chr8_21914018_21914169 | 0.03 |
DMTN |
dematin actin binding protein |
323 |
0.82 |
| chr6_15297900_15298051 | 0.03 |
ENSG00000201367 |
. |
17176 |
0.18 |
| chr15_58767167_58767318 | 0.03 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
1130 |
0.49 |
| chr11_47586558_47587254 | 0.03 |
PTPMT1 |
protein tyrosine phosphatase, mitochondrial 1 |
76 |
0.59 |
| chr1_205343234_205343399 | 0.03 |
LEMD1-AS1 |
LEMD1 antisense RNA 1 |
936 |
0.53 |
| chr4_156681206_156681472 | 0.03 |
GUCY1B3 |
guanylate cyclase 1, soluble, beta 3 |
728 |
0.72 |
| chr9_34047108_34047259 | 0.03 |
UBAP2 |
ubiquitin associated protein 2 |
1700 |
0.27 |
| chr21_36594684_36594936 | 0.03 |
RUNX1 |
runt-related transcription factor 1 |
173169 |
0.04 |
| chr19_2933350_2933631 | 0.03 |
AC006277.2 |
|
6685 |
0.12 |
| chr7_50321625_50321823 | 0.03 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
22600 |
0.25 |
| chr4_70108820_70108971 | 0.03 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
28446 |
0.16 |
| chr19_58772914_58773185 | 0.03 |
CTD-3138B18.6 |
|
4952 |
0.11 |
| chr8_1993393_1993636 | 0.03 |
MYOM2 |
myomesin 2 |
330 |
0.93 |
| chr19_58370756_58370907 | 0.03 |
ZNF587 |
zinc finger protein 587 |
3494 |
0.13 |
| chr18_42044646_42044797 | 0.03 |
SETBP1 |
SET binding protein 1 |
215417 |
0.02 |
| chr19_20117293_20117444 | 0.03 |
ZNF682 |
zinc finger protein 682 |
703 |
0.66 |
| chr19_38229308_38229579 | 0.03 |
ZNF607 |
zinc finger protein 607 |
18752 |
0.15 |
| chr6_142699302_142699594 | 0.03 |
GPR126 |
G protein-coupled receptor 126 |
12001 |
0.29 |
| chr19_35448920_35449083 | 0.03 |
ZNF792 |
zinc finger protein 792 |
2606 |
0.2 |
| chr14_65184455_65184704 | 0.03 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
2158 |
0.36 |
| chr19_12541727_12541878 | 0.03 |
CTD-3105H18.16 |
Uncharacterized protein |
10094 |
0.11 |
| chr6_28504701_28505374 | 0.03 |
GPX5 |
glutathione peroxidase 5 (epididymal androgen-related protein) |
11248 |
0.17 |
| chr9_124414074_124414751 | 0.03 |
DAB2IP |
DAB2 interacting protein |
539 |
0.84 |
| chr4_149137803_149137954 | 0.03 |
RP11-76G10.1 |
|
70256 |
0.13 |
| chr1_15251371_15251898 | 0.03 |
KAZN |
kazrin, periplakin interacting protein |
1008 |
0.7 |
| chrX_19687578_19687729 | 0.03 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
827 |
0.76 |
| chr6_7469002_7469180 | 0.03 |
RP3-512B11.3 |
|
72480 |
0.09 |
| chr5_80716043_80716194 | 0.03 |
ACOT12 |
acyl-CoA thioesterase 12 |
26120 |
0.23 |
| chr3_14853690_14853962 | 0.03 |
FGD5 |
FYVE, RhoGEF and PH domain containing 5 |
6643 |
0.24 |
| chr15_62402344_62402495 | 0.03 |
C2CD4A |
C2 calcium-dependent domain containing 4A |
43243 |
0.13 |
| chr10_95333061_95333288 | 0.03 |
FFAR4 |
free fatty acid receptor 4 |
6735 |
0.17 |
| chr1_247201400_247201551 | 0.03 |
ZNF695 |
zinc finger protein 695 |
30080 |
0.15 |
| chr2_144499859_144500010 | 0.03 |
RP11-434H14.1 |
|
1071 |
0.59 |
| chr4_184826206_184826847 | 0.03 |
STOX2 |
storkhead box 2 |
17 |
0.99 |
| chr2_95880533_95880684 | 0.03 |
ZNF2 |
zinc finger protein 2 |
49046 |
0.12 |
| chr6_113953461_113953992 | 0.03 |
ENSG00000266650 |
. |
29609 |
0.21 |
| chr5_92924582_92924758 | 0.03 |
ENSG00000237187 |
. |
3316 |
0.26 |
| chr16_72882732_72883241 | 0.03 |
ENSG00000251868 |
. |
27095 |
0.17 |
| chr6_90796198_90796349 | 0.03 |
ENSG00000222078 |
. |
85048 |
0.09 |
| chr14_74222749_74222900 | 0.03 |
ELMSAN1 |
ELM2 and Myb/SANT-like domain containing 1 |
1105 |
0.36 |
| chr12_49392080_49393040 | 0.03 |
RP11-386G11.5 |
|
379 |
0.49 |
| chr2_220251514_220252585 | 0.03 |
DNPEP |
aspartyl aminopeptidase |
19 |
0.94 |
| chr19_58861430_58861581 | 0.03 |
A1BG-AS1 |
A1BG antisense RNA 1 |
1831 |
0.13 |
| chr9_112889621_112889772 | 0.03 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
1915 |
0.45 |
| chr7_36195246_36195841 | 0.03 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
2666 |
0.32 |
| chr4_139941093_139941244 | 0.03 |
ENSG00000264953 |
. |
2435 |
0.22 |
| chr9_12775957_12776199 | 0.03 |
LURAP1L |
leucine rich adaptor protein 1-like |
1058 |
0.54 |
| chr20_55363223_55363386 | 0.03 |
ENSG00000207158 |
. |
2161 |
0.37 |
| chr5_159738704_159739455 | 0.03 |
CCNJL |
cyclin J-like |
404 |
0.84 |
| chr10_110225810_110226152 | 0.03 |
ENSG00000222436 |
. |
475087 |
0.01 |
| chr9_129467466_129467647 | 0.03 |
ENSG00000266403 |
. |
13056 |
0.21 |
| chr1_15575745_15575896 | 0.03 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
2044 |
0.36 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |