Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
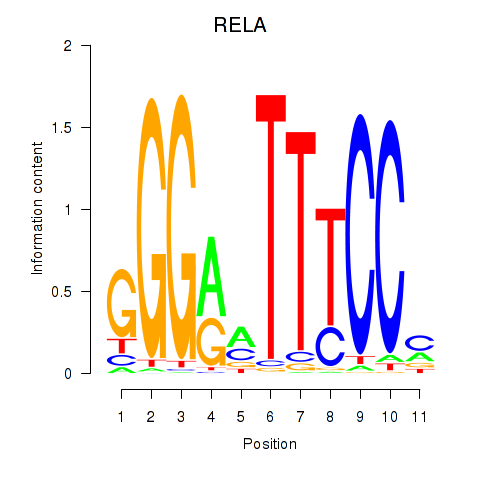
Results for RELA
Z-value: 0.63
Transcription factors associated with RELA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RELA
|
ENSG00000173039.14 | RELA proto-oncogene, NF-kB subunit |
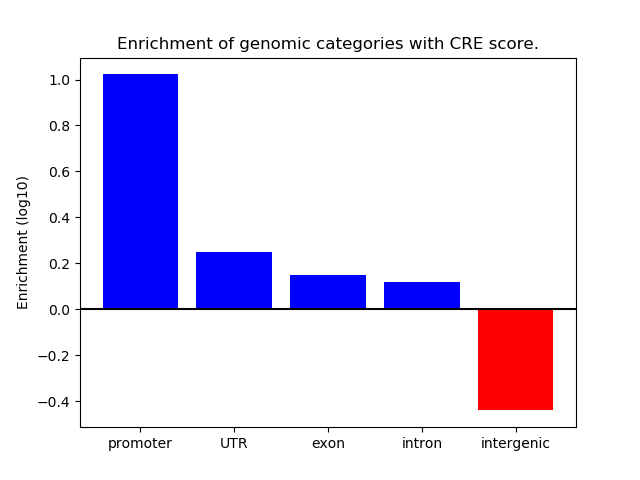
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_65429318_65430091 | RELA | 195 | 0.869044 | -0.42 | 2.6e-01 | Click! |
| chr11_65430100_65430433 | RELA | 2 | 0.946657 | -0.23 | 5.5e-01 | Click! |
| chr11_65430663_65431111 | RELA | 322 | 0.764562 | -0.18 | 6.5e-01 | Click! |
| chr11_65431137_65431406 | RELA | 706 | 0.461078 | 0.07 | 8.7e-01 | Click! |
Activity of the RELA motif across conditions
Conditions sorted by the z-value of the RELA motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
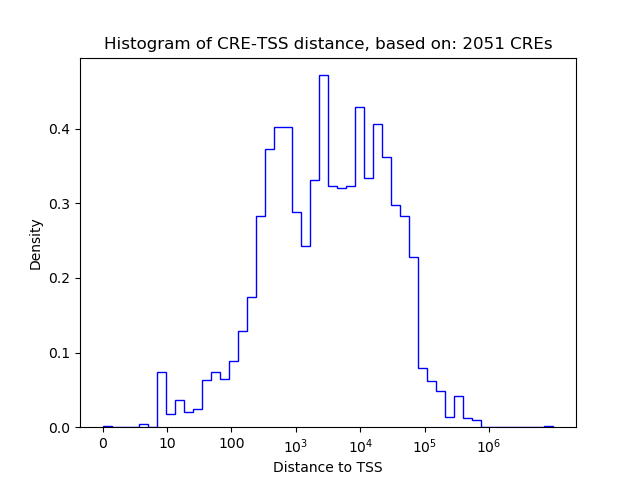
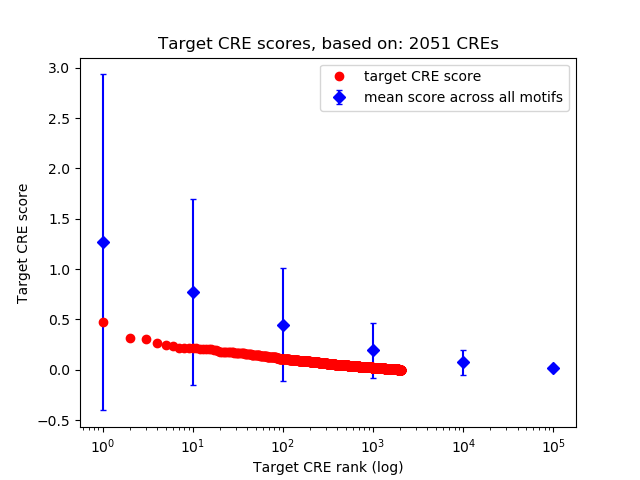
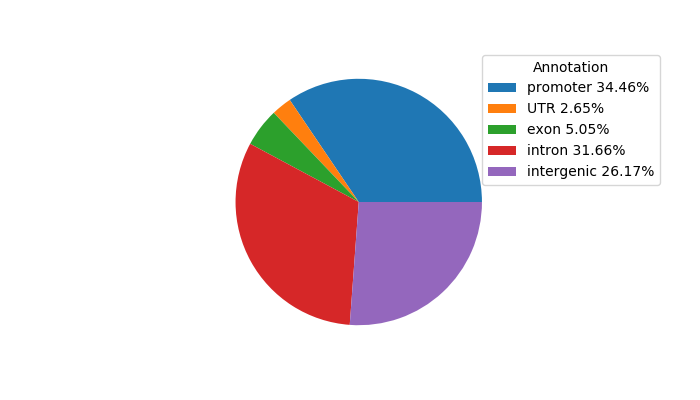
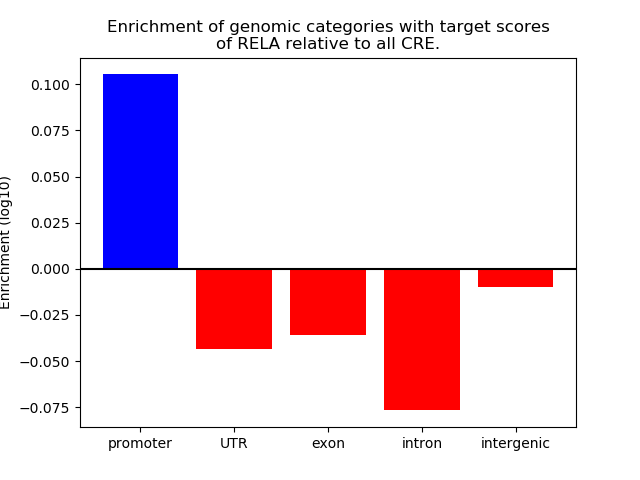
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_8000859_8001010 | 0.47 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
8 |
0.98 |
| chr22_30475055_30475206 | 0.32 |
HORMAD2 |
HORMA domain containing 2 |
1033 |
0.38 |
| chrX_40429955_40430133 | 0.30 |
ATP6AP2 |
ATPase, H+ transporting, lysosomal accessory protein 2 |
10102 |
0.21 |
| chr20_4795121_4795462 | 0.26 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
478 |
0.82 |
| chr12_42958835_42958986 | 0.24 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
24568 |
0.21 |
| chr14_94425760_94425911 | 0.24 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
2068 |
0.26 |
| chr7_128578118_128579002 | 0.22 |
IRF5 |
interferon regulatory factor 5 |
289 |
0.89 |
| chr6_20202498_20202689 | 0.22 |
RP11-239H6.2 |
|
9725 |
0.23 |
| chr15_69083910_69084061 | 0.22 |
ENSG00000265195 |
. |
10279 |
0.23 |
| chr4_100738641_100738957 | 0.21 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
796 |
0.72 |
| chrX_53741093_53741244 | 0.21 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
27495 |
0.23 |
| chr20_5296763_5297139 | 0.21 |
PROKR2 |
prokineticin receptor 2 |
427 |
0.87 |
| chr10_65021596_65021747 | 0.21 |
JMJD1C |
jumonji domain containing 1C |
7155 |
0.29 |
| chr2_38265076_38265999 | 0.21 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
2053 |
0.33 |
| chr2_214108249_214108400 | 0.21 |
SPAG16 |
sperm associated antigen 16 |
40789 |
0.21 |
| chr20_50141923_50142074 | 0.21 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
17260 |
0.25 |
| chr17_47572677_47573321 | 0.20 |
NGFR |
nerve growth factor receptor |
344 |
0.86 |
| chr3_71833354_71833608 | 0.19 |
PROK2 |
prokineticin 2 |
731 |
0.68 |
| chr2_113932280_113932569 | 0.19 |
AC016683.5 |
|
490 |
0.59 |
| chr14_66694537_66694688 | 0.18 |
ENSG00000200860 |
. |
50259 |
0.19 |
| chr5_150473764_150473973 | 0.18 |
TNIP1 |
TNFAIP3 interacting protein 1 |
730 |
0.68 |
| chr8_126964027_126964178 | 0.18 |
ENSG00000206695 |
. |
50907 |
0.19 |
| chr9_123690633_123691433 | 0.18 |
TRAF1 |
TNF receptor-associated factor 1 |
14 |
0.98 |
| chrX_9979270_9979421 | 0.18 |
WWC3 |
WWC family member 3 |
4257 |
0.22 |
| chr5_49962630_49963750 | 0.18 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
201 |
0.97 |
| chr2_219745053_219745526 | 0.18 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
204 |
0.9 |
| chr15_45409056_45409207 | 0.17 |
DUOX2 |
dual oxidase 2 |
2589 |
0.14 |
| chr13_46739755_46739906 | 0.17 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
2824 |
0.22 |
| chr1_9688741_9688898 | 0.17 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
22971 |
0.15 |
| chr6_138028959_138029110 | 0.17 |
ENSG00000216097 |
. |
9051 |
0.27 |
| chr3_142800787_142800938 | 0.17 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
37311 |
0.15 |
| chr15_31638589_31638828 | 0.17 |
KLF13 |
Kruppel-like factor 13 |
6572 |
0.32 |
| chr16_28996496_28996848 | 0.16 |
LAT |
linker for activation of T cells |
11 |
0.94 |
| chr2_164608573_164608724 | 0.16 |
FIGN |
fidgetin |
16126 |
0.31 |
| chr17_72735282_72735631 | 0.16 |
RAB37 |
RAB37, member RAS oncogene family |
2085 |
0.18 |
| chr14_22994691_22994955 | 0.16 |
TRAJ15 |
T cell receptor alpha joining 15 |
3757 |
0.14 |
| chr7_30766706_30766866 | 0.16 |
INMT |
indolethylamine N-methyltransferase |
24965 |
0.16 |
| chr4_105982370_105982725 | 0.16 |
ENSG00000252136 |
. |
44597 |
0.16 |
| chr12_62996165_62996975 | 0.16 |
RP11-631N16.2 |
|
38 |
0.9 |
| chr1_112159044_112159195 | 0.16 |
RAP1A |
RAP1A, member of RAS oncogene family |
3280 |
0.23 |
| chr2_10548224_10548465 | 0.16 |
HPCAL1 |
hippocalcin-like 1 |
11803 |
0.19 |
| chr19_54672117_54672268 | 0.16 |
TMC4 |
transmembrane channel-like 4 |
4255 |
0.08 |
| chr2_10169297_10169448 | 0.16 |
KLF11 |
Kruppel-like factor 11 |
13604 |
0.13 |
| chr19_45271245_45271464 | 0.15 |
ENSG00000253027 |
. |
3147 |
0.15 |
| chr1_93443435_93443586 | 0.15 |
ENSG00000239710 |
. |
2830 |
0.16 |
| chr3_195913428_195913625 | 0.15 |
ENSG00000222335 |
. |
15694 |
0.13 |
| chr16_68798920_68799109 | 0.15 |
ENSG00000200558 |
. |
22631 |
0.14 |
| chr5_1105065_1105874 | 0.15 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
6681 |
0.19 |
| chr8_37751638_37751789 | 0.15 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
5259 |
0.15 |
| chr15_31555832_31555983 | 0.15 |
KLF13 |
Kruppel-like factor 13 |
63151 |
0.13 |
| chr9_123688442_123688941 | 0.15 |
TRAF1 |
TNF receptor-associated factor 1 |
2356 |
0.33 |
| chr9_139927070_139927409 | 0.14 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
223 |
0.78 |
| chr6_3876229_3876380 | 0.14 |
FAM50B |
family with sequence similarity 50, member B |
26682 |
0.18 |
| chr13_74807358_74807666 | 0.14 |
ENSG00000206617 |
. |
55839 |
0.16 |
| chr17_46187551_46187761 | 0.14 |
SNX11 |
sorting nexin 11 |
1655 |
0.23 |
| chr17_38024211_38024362 | 0.14 |
ZPBP2 |
zona pellucida binding protein 2 |
131 |
0.95 |
| chr17_66286056_66286766 | 0.14 |
SLC16A6 |
solute carrier family 16, member 6 |
846 |
0.47 |
| chr16_89690039_89690304 | 0.14 |
DPEP1 |
dipeptidase 1 (renal) |
3171 |
0.14 |
| chrX_123373984_123374135 | 0.14 |
ENSG00000252693 |
. |
42466 |
0.19 |
| chr10_90753077_90753312 | 0.14 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
2047 |
0.24 |
| chr7_36640832_36640983 | 0.14 |
AOAH-IT1 |
AOAH intronic transcript 1 (non-protein coding) |
1181 |
0.5 |
| chr1_150737104_150737638 | 0.14 |
CTSS |
cathepsin S |
897 |
0.5 |
| chr21_30464791_30465071 | 0.13 |
MAP3K7CL |
MAP3K7 C-terminal like |
356 |
0.83 |
| chr5_134879321_134879990 | 0.13 |
NEUROG1 |
neurogenin 1 |
8016 |
0.18 |
| chr1_147229260_147229411 | 0.13 |
RP11-433J22.2 |
|
91 |
0.97 |
| chr3_131105945_131106096 | 0.13 |
NUDT16 |
nudix (nucleoside diphosphate linked moiety X)-type motif 16 |
5391 |
0.18 |
| chr20_34674700_34674851 | 0.13 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
4651 |
0.22 |
| chr15_86127194_86127729 | 0.13 |
RP11-815J21.2 |
|
4052 |
0.21 |
| chr11_121323015_121324255 | 0.13 |
RP11-730K11.1 |
|
7 |
0.95 |
| chr21_40275169_40275320 | 0.13 |
ENSG00000272015 |
. |
8535 |
0.29 |
| chr1_31227757_31227908 | 0.13 |
LAPTM5 |
lysosomal protein transmembrane 5 |
2835 |
0.23 |
| chr1_199997854_199998005 | 0.13 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
918 |
0.55 |
| chr14_90798758_90798909 | 0.13 |
NRDE2 |
NRDE-2, necessary for RNA interference, domain containing |
352 |
0.89 |
| chr7_74057672_74058172 | 0.13 |
GTF2I |
general transcription factor IIi |
14089 |
0.22 |
| chr1_166459489_166459697 | 0.13 |
FAM78B |
family with sequence similarity 78, member B |
323387 |
0.01 |
| chr9_134143747_134144006 | 0.13 |
FAM78A |
family with sequence similarity 78, member A |
2004 |
0.31 |
| chr12_120679359_120679510 | 0.12 |
PXN |
paxillin |
8530 |
0.12 |
| chrX_128915626_128915992 | 0.12 |
SASH3 |
SAM and SH3 domain containing 3 |
1849 |
0.35 |
| chr11_27435035_27435186 | 0.12 |
CCDC34 |
coiled-coil domain containing 34 |
49695 |
0.12 |
| chr17_38480138_38480579 | 0.12 |
RARA |
retinoic acid receptor, alpha |
5821 |
0.12 |
| chr14_77499378_77500437 | 0.12 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
4873 |
0.22 |
| chr11_114217713_114217864 | 0.12 |
RP11-64D24.2 |
|
9505 |
0.18 |
| chr15_81586254_81586487 | 0.12 |
IL16 |
interleukin 16 |
2884 |
0.28 |
| chr20_47441337_47441845 | 0.12 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
2829 |
0.36 |
| chr10_6243545_6244657 | 0.12 |
RP11-414H17.5 |
|
555 |
0.52 |
| chr12_22695256_22695407 | 0.12 |
C2CD5 |
C2 calcium-dependent domain containing 5 |
1856 |
0.46 |
| chrX_48793999_48794379 | 0.12 |
PIM2 |
pim-2 oncogene |
17888 |
0.08 |
| chrX_41301243_41301541 | 0.12 |
NYX |
nyctalopin |
5295 |
0.25 |
| chr7_26333293_26333815 | 0.11 |
SNX10 |
sorting nexin 10 |
880 |
0.67 |
| chr1_65430726_65432146 | 0.11 |
JAK1 |
Janus kinase 1 |
751 |
0.73 |
| chr6_106808594_106809355 | 0.11 |
RP11-404H14.1 |
|
1382 |
0.42 |
| chr9_134604865_134605016 | 0.11 |
ENSG00000240853 |
. |
4086 |
0.24 |
| chr8_144512945_144513817 | 0.11 |
MAFA |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
805 |
0.48 |
| chr21_39867301_39867487 | 0.11 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
2951 |
0.4 |
| chr4_17809379_17809707 | 0.11 |
DCAF16 |
DDB1 and CUL4 associated factor 16 |
2838 |
0.25 |
| chr6_1014051_1014202 | 0.11 |
AL033381.1 |
Uncharacterized protein; cDNA FLJ34594 fis, clone KIDNE2009109 |
66038 |
0.14 |
| chr22_42815517_42815668 | 0.11 |
NFAM1 |
NFAT activating protein with ITAM motif 1 |
12809 |
0.19 |
| chr6_132113440_132113591 | 0.11 |
ENSG00000266807 |
. |
203 |
0.94 |
| chr9_132650035_132650502 | 0.11 |
FNBP1 |
formin binding protein 1 |
31321 |
0.13 |
| chr3_16156075_16156226 | 0.11 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
60006 |
0.14 |
| chr15_41234314_41234975 | 0.11 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
10516 |
0.12 |
| chr2_33537454_33537605 | 0.11 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
10932 |
0.2 |
| chr10_3848477_3848747 | 0.11 |
KLF6 |
Kruppel-like factor 6 |
21139 |
0.22 |
| chr18_59519452_59519603 | 0.11 |
RNF152 |
ring finger protein 152 |
41465 |
0.22 |
| chr17_67322489_67323211 | 0.11 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
381 |
0.89 |
| chr11_69457554_69457705 | 0.11 |
CCND1 |
cyclin D1 |
1655 |
0.41 |
| chr1_226922169_226922483 | 0.11 |
ITPKB |
inositol-trisphosphate 3-kinase B |
2833 |
0.32 |
| chr12_111842372_111842523 | 0.11 |
SH2B3 |
SH2B adaptor protein 3 |
1305 |
0.45 |
| chr1_37945353_37945703 | 0.11 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
1763 |
0.28 |
| chr19_50400390_50400724 | 0.11 |
IL4I1 |
interleukin 4 induced 1 |
345 |
0.67 |
| chr1_143743180_143744039 | 0.11 |
RP6-206I17.4 |
|
23905 |
0.15 |
| chr6_72155810_72156092 | 0.11 |
ENSG00000207827 |
. |
42627 |
0.16 |
| chr12_111115977_111116128 | 0.11 |
HVCN1 |
hydrogen voltage-gated channel 1 |
10462 |
0.16 |
| chr15_42349005_42349667 | 0.11 |
PLA2G4E |
phospholipase A2, group IVE |
5948 |
0.18 |
| chr18_59619511_59619683 | 0.10 |
RNF152 |
ring finger protein 152 |
58133 |
0.16 |
| chr19_10341031_10341948 | 0.10 |
ENSG00000264266 |
. |
400 |
0.34 |
| chr15_91807157_91807528 | 0.10 |
SV2B |
synaptic vesicle glycoprotein 2B |
38242 |
0.2 |
| chr8_142138821_142140144 | 0.10 |
RP11-809O17.1 |
|
578 |
0.46 |
| chr2_219744680_219744914 | 0.10 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
288 |
0.85 |
| chr22_22005684_22006266 | 0.10 |
ENSG00000212102 |
. |
1295 |
0.23 |
| chr8_59717634_59717850 | 0.10 |
ENSG00000201231 |
. |
9369 |
0.29 |
| chr6_37592383_37592534 | 0.10 |
MDGA1 |
MAM domain containing glycosylphosphatidylinositol anchor 1 |
21692 |
0.22 |
| chr17_56230401_56230552 | 0.10 |
OR4D1 |
olfactory receptor, family 4, subfamily D, member 1 |
2018 |
0.26 |
| chr6_75458992_75459143 | 0.10 |
ENSG00000264884 |
. |
29508 |
0.26 |
| chr17_7152312_7152463 | 0.10 |
CTDNEP1 |
CTD nuclear envelope phosphatase 1 |
1729 |
0.12 |
| chr10_91093714_91093865 | 0.10 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
1548 |
0.3 |
| chr7_139930052_139930405 | 0.10 |
ENSG00000199283 |
. |
11442 |
0.17 |
| chr18_42306038_42306201 | 0.10 |
SETBP1 |
SET binding protein 1 |
28990 |
0.26 |
| chr12_56881156_56881307 | 0.10 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
671 |
0.58 |
| chr14_93427433_93427584 | 0.10 |
CHGA |
chromogranin A (parathyroid secretory protein 1) |
37987 |
0.18 |
| chr1_154300198_154300512 | 0.10 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
79 |
0.94 |
| chr19_1074414_1074782 | 0.10 |
HMHA1 |
histocompatibility (minor) HA-1 |
2844 |
0.13 |
| chr18_72912801_72912952 | 0.10 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
4592 |
0.28 |
| chr19_46283149_46283300 | 0.10 |
DMPK |
dystrophia myotonica-protein kinase |
575 |
0.52 |
| chr1_247354543_247354824 | 0.10 |
MIR3916 |
microRNA 3916 |
8815 |
0.14 |
| chr9_92218215_92218814 | 0.10 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
1414 |
0.56 |
| chr21_45564255_45564539 | 0.10 |
C21orf33 |
chromosome 21 open reading frame 33 |
8396 |
0.15 |
| chr20_43597999_43598419 | 0.10 |
STK4 |
serine/threonine kinase 4 |
3042 |
0.19 |
| chr10_63595750_63596410 | 0.10 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
64979 |
0.13 |
| chr4_113205771_113206741 | 0.10 |
TIFA |
TRAF-interacting protein with forkhead-associated domain |
803 |
0.58 |
| chr2_145116612_145116763 | 0.10 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
26604 |
0.24 |
| chr7_4817960_4818111 | 0.10 |
AP5Z1 |
adaptor-related protein complex 5, zeta 1 subunit |
2771 |
0.22 |
| chr1_51442471_51443647 | 0.10 |
CDKN2C |
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
7443 |
0.21 |
| chr2_47796814_47797322 | 0.10 |
KCNK12 |
potassium channel, subfamily K, member 12 |
1010 |
0.5 |
| chr2_106363775_106364396 | 0.10 |
NCK2 |
NCK adaptor protein 2 |
1897 |
0.48 |
| chr1_167599991_167600483 | 0.10 |
RP3-455J7.4 |
|
326 |
0.69 |
| chr19_38908265_38908571 | 0.09 |
RASGRP4 |
RAS guanyl releasing protein 4 |
8384 |
0.09 |
| chr9_117145261_117145418 | 0.09 |
AKNA |
AT-hook transcription factor |
4904 |
0.23 |
| chr2_192058910_192059074 | 0.09 |
STAT4 |
signal transducer and activator of transcription 4 |
42670 |
0.15 |
| chr5_149461865_149462126 | 0.09 |
CSF1R |
colony stimulating factor 1 receptor |
3995 |
0.18 |
| chr18_2656091_2657266 | 0.09 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
792 |
0.58 |
| chr8_28205045_28205788 | 0.09 |
ZNF395 |
zinc finger protein 395 |
3667 |
0.19 |
| chr19_49839028_49839359 | 0.09 |
CD37 |
CD37 molecule |
472 |
0.63 |
| chr10_79050334_79050485 | 0.09 |
RP11-328K22.1 |
|
23290 |
0.23 |
| chr22_33278140_33278291 | 0.09 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
80528 |
0.1 |
| chr20_47884918_47885071 | 0.09 |
ZNFX1 |
zinc finger, NFX1-type containing 1 |
2766 |
0.18 |
| chr20_3888044_3888195 | 0.09 |
ENSG00000199024 |
. |
10022 |
0.14 |
| chr1_36816210_36816695 | 0.09 |
STK40 |
serine/threonine kinase 40 |
10489 |
0.13 |
| chr2_234293590_234293937 | 0.09 |
DGKD |
diacylglycerol kinase, delta 130kDa |
828 |
0.57 |
| chr6_144017218_144017369 | 0.09 |
PHACTR2 |
phosphatase and actin regulator 2 |
18088 |
0.25 |
| chr4_152331308_152331459 | 0.09 |
FAM160A1 |
family with sequence similarity 160, member A1 |
879 |
0.7 |
| chr12_806850_807001 | 0.09 |
NINJ2 |
ninjurin 2 |
33980 |
0.14 |
| chr19_39106930_39107372 | 0.09 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
1413 |
0.27 |
| chr2_201987300_201987657 | 0.09 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
278 |
0.87 |
| chr20_1309605_1309756 | 0.09 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
203 |
0.91 |
| chr6_15309152_15309303 | 0.09 |
ENSG00000201367 |
. |
5924 |
0.22 |
| chr11_71826248_71826399 | 0.09 |
ANAPC15 |
anaphase promoting complex subunit 15 |
2497 |
0.13 |
| chr9_114792415_114792566 | 0.09 |
RP11-4O1.2 |
|
6100 |
0.22 |
| chr17_37793084_37793352 | 0.09 |
STARD3 |
StAR-related lipid transfer (START) domain containing 3 |
100 |
0.94 |
| chr1_27070439_27070710 | 0.09 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
14703 |
0.13 |
| chr7_29283709_29283860 | 0.09 |
AC004593.3 |
|
35198 |
0.16 |
| chrX_39473449_39473600 | 0.09 |
ENSG00000263730 |
. |
46946 |
0.2 |
| chrX_30594869_30596024 | 0.09 |
CXorf21 |
chromosome X open reading frame 21 |
515 |
0.84 |
| chr15_45027870_45028221 | 0.09 |
TRIM69 |
tripartite motif containing 69 |
515 |
0.75 |
| chr1_203259093_203259633 | 0.09 |
BTG2 |
BTG family, member 2 |
15301 |
0.16 |
| chr9_37036694_37037719 | 0.09 |
PAX5 |
paired box 5 |
3103 |
0.27 |
| chr7_74194192_74194343 | 0.09 |
NCF1 |
neutrophil cytosolic factor 1 |
2654 |
0.28 |
| chr20_11435441_11435592 | 0.09 |
ENSG00000222281 |
. |
364223 |
0.01 |
| chr11_14468343_14468494 | 0.09 |
ENSG00000251991 |
. |
32020 |
0.18 |
| chr1_182583862_182584759 | 0.09 |
RGS16 |
regulator of G-protein signaling 16 |
10767 |
0.21 |
| chr19_35940283_35940434 | 0.09 |
FFAR2 |
free fatty acid receptor 2 |
259 |
0.87 |
| chr9_72725675_72725941 | 0.09 |
MAMDC2-AS1 |
MAMDC2 antisense RNA 1 |
2968 |
0.33 |
| chr3_33800152_33800303 | 0.08 |
PDCD6IP |
programmed cell death 6 interacting protein |
39617 |
0.18 |
| chrX_76776329_76776480 | 0.08 |
FGF16 |
fibroblast growth factor 16 |
66756 |
0.13 |
| chr16_16119769_16119920 | 0.08 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
16131 |
0.23 |
| chr12_70132575_70133879 | 0.08 |
RAB3IP |
RAB3A interacting protein |
47 |
0.95 |
| chr16_3095827_3096553 | 0.08 |
MMP25 |
matrix metallopeptidase 25 |
492 |
0.51 |
| chr2_28904176_28904327 | 0.08 |
ENSG00000222232 |
. |
2591 |
0.25 |
| chr20_25037809_25038550 | 0.08 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
639 |
0.73 |
| chr9_110187375_110187686 | 0.08 |
ENSG00000263459 |
. |
8360 |
0.24 |
| chr3_9745623_9746706 | 0.08 |
CPNE9 |
copine family member IX |
654 |
0.62 |
| chr7_5183512_5184010 | 0.08 |
ZNF890P |
zinc finger protein 890, pseudogene |
16335 |
0.17 |
| chr7_44679217_44679482 | 0.08 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
15288 |
0.18 |
| chr15_91373266_91373417 | 0.08 |
CTD-3094K11.1 |
|
9613 |
0.13 |
| chrX_48794595_48794842 | 0.08 |
PIM2 |
pim-2 oncogene |
18417 |
0.08 |
| chr20_35463892_35464165 | 0.08 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
19344 |
0.15 |
| chr13_20175736_20176974 | 0.08 |
MPHOSPH8 |
M-phase phosphoprotein 8 |
31433 |
0.18 |
| chr11_35325325_35325476 | 0.08 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
2325 |
0.31 |
| chr2_43452698_43453520 | 0.08 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
639 |
0.76 |
| chr4_103421804_103422645 | 0.08 |
AF213884.2 |
|
252 |
0.5 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0052167 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:1903521 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0043303 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.2 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |