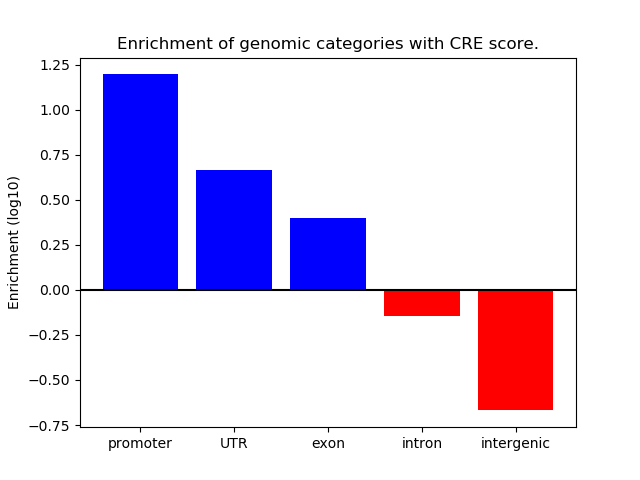
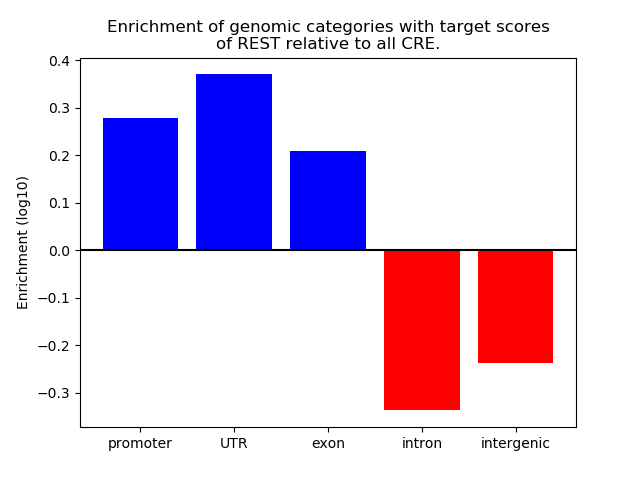
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
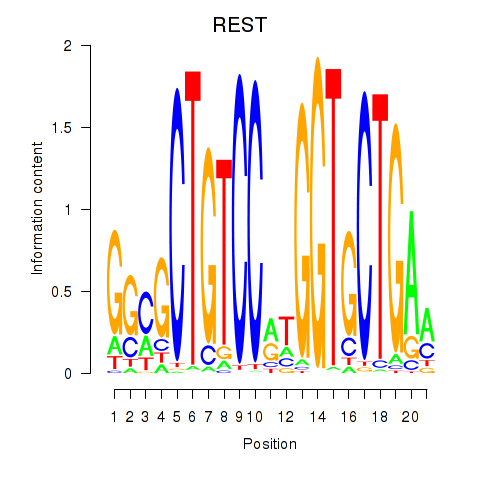
Results for REST
Z-value: 0.41
Transcription factors associated with REST
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
REST
|
ENSG00000084093.11 | RE1 silencing transcription factor |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_57773669_57773922 | REST | 280 | 0.912776 | -0.55 | 1.3e-01 | Click! |
| chr4_57737524_57737675 | REST | 36476 | 0.124522 | 0.54 | 1.3e-01 | Click! |
| chr4_57773435_57773624 | REST | 546 | 0.775427 | -0.50 | 1.7e-01 | Click! |
| chr4_57775517_57776061 | REST | 1714 | 0.365889 | -0.31 | 4.1e-01 | Click! |
| chr4_57737703_57737910 | REST | 36269 | 0.125070 | 0.31 | 4.2e-01 | Click! |
Activity of the REST motif across conditions
Conditions sorted by the z-value of the REST motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
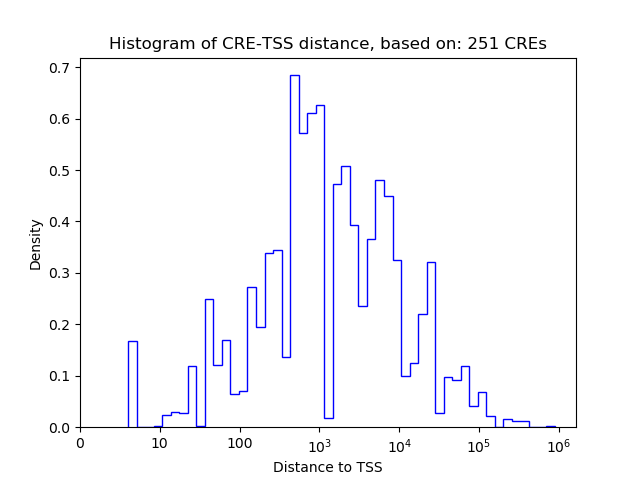
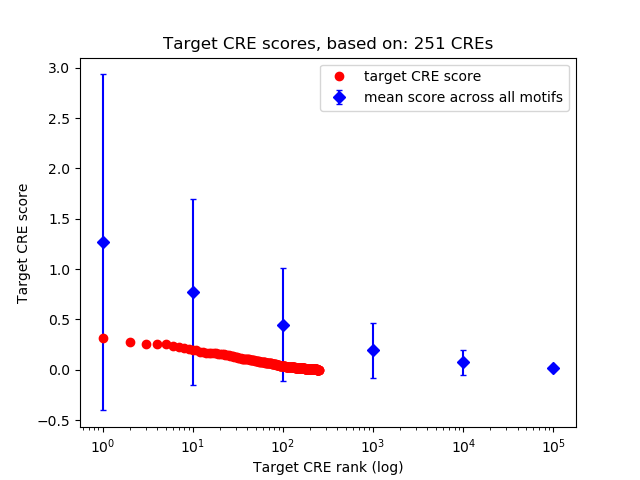
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr21_42218839_42219216 | 0.31 |
DSCAM |
Down syndrome cell adhesion molecule |
38 |
0.99 |
| chr3_9745623_9746706 | 0.28 |
CPNE9 |
copine family member IX |
654 |
0.62 |
| chr1_31331684_31331943 | 0.26 |
RP1-65J11.5 |
|
808 |
0.58 |
| chr1_1899902_1900053 | 0.25 |
C1orf222 |
chromosome 1 open reading frame 222 |
2012 |
0.25 |
| chr10_50606076_50606780 | 0.25 |
DRGX |
dorsal root ganglia homeobox |
2931 |
0.27 |
| chr14_58332211_58332467 | 0.24 |
SLC35F4 |
solute carrier family 35, member F4 |
441 |
0.87 |
| chr2_51259112_51259522 | 0.23 |
NRXN1 |
neurexin 1 |
3 |
0.99 |
| chr19_10806604_10806755 | 0.22 |
QTRT1 |
queuine tRNA-ribosyltransferase 1 |
5448 |
0.12 |
| chr20_33814974_33815173 | 0.21 |
MMP24 |
matrix metallopeptidase 24 (membrane-inserted) |
616 |
0.6 |
| chr1_20916301_20916452 | 0.20 |
CDA |
cytidine deaminase |
935 |
0.55 |
| chr1_43201434_43201585 | 0.19 |
CLDN19 |
claudin 19 |
4302 |
0.16 |
| chr16_2510179_2510910 | 0.17 |
C16orf59 |
chromosome 16 open reading frame 59 |
67 |
0.92 |
| chr10_89419474_89420355 | 0.17 |
RP11-57C13.3 |
|
154 |
0.8 |
| chr5_6687085_6687491 | 0.17 |
PAPD7 |
PAP associated domain containing 7 |
27430 |
0.2 |
| chr7_948384_949423 | 0.17 |
ADAP1 |
ArfGAP with dual PH domains 1 |
4189 |
0.17 |
| chr2_47798093_47798638 | 0.17 |
KCNK12 |
potassium channel, subfamily K, member 12 |
287 |
0.86 |
| chr6_43252045_43252196 | 0.17 |
SLC22A7 |
solute carrier family 22 (organic anion transporter), member 7 |
13616 |
0.13 |
| chr16_30662970_30663539 | 0.17 |
PRR14 |
proline rich 14 |
743 |
0.43 |
| chr15_72612091_72612308 | 0.16 |
CELF6 |
CUGBP, Elav-like family member 6 |
24 |
0.97 |
| chr19_11592760_11593066 | 0.16 |
ELAVL3 |
ELAV like neuron-specific RNA binding protein 3 |
1065 |
0.31 |
| chr3_131245838_131246060 | 0.16 |
RP11-517B11.4 |
|
9274 |
0.2 |
| chr8_57358039_57358590 | 0.15 |
RP11-17A4.2 |
|
52 |
0.69 |
| chr1_17896482_17896633 | 0.15 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
10491 |
0.26 |
| chr8_145752569_145752720 | 0.15 |
LRRC24 |
leucine rich repeat containing 24 |
228 |
0.77 |
| chr19_55728419_55728773 | 0.14 |
PTPRH |
protein tyrosine phosphatase, receptor type, H |
7772 |
0.08 |
| chr16_89992729_89992956 | 0.14 |
TUBB3 |
Tubulin beta-3 chain |
3068 |
0.14 |
| chr1_155973189_155973340 | 0.14 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
3597 |
0.12 |
| chr13_114540399_114540680 | 0.14 |
GAS6 |
growth arrest-specific 6 |
1522 |
0.45 |
| chr5_54516767_54517029 | 0.13 |
MCIDAS |
multiciliate differentiation and DNA synthesis associated cell cycle protein |
6245 |
0.13 |
| chr11_101453394_101453891 | 0.12 |
TRPC6 |
transient receptor potential cation channel, subfamily C, member 6 |
592 |
0.84 |
| chr20_43273137_43273288 | 0.12 |
ADA |
adenosine deaminase |
7120 |
0.14 |
| chr11_89785091_89785242 | 0.12 |
TRIM49C |
tripartite motif containing 49C |
20892 |
0.22 |
| chr19_8305457_8305834 | 0.12 |
CTD-3020H12.4 |
|
1113 |
0.43 |
| chr20_62528184_62528335 | 0.11 |
DNAJC5 |
DnaJ (Hsp40) homolog, subfamily C, member 5 |
1724 |
0.16 |
| chr16_68269517_68269668 | 0.11 |
ESRP2 |
epithelial splicing regulatory protein 2 |
443 |
0.6 |
| chr14_24906611_24906867 | 0.11 |
SDR39U1 |
short chain dehydrogenase/reductase family 39U, member 1 |
3518 |
0.13 |
| chr14_75926184_75926335 | 0.11 |
JDP2 |
Jun dimerization protein 2 |
27422 |
0.16 |
| chr3_49169788_49170138 | 0.11 |
LAMB2 |
laminin, beta 2 (laminin S) |
490 |
0.65 |
| chr5_168727419_168727744 | 0.11 |
SLIT3 |
slit homolog 3 (Drosophila) |
132 |
0.98 |
| chr7_87258351_87258502 | 0.10 |
RUNDC3B |
RUN domain containing 3B |
495 |
0.81 |
| chr6_36975031_36975182 | 0.10 |
FGD2 |
FYVE, RhoGEF and PH domain containing 2 |
1683 |
0.34 |
| chr5_32232128_32232279 | 0.10 |
ENSG00000199731 |
. |
2778 |
0.29 |
| chr3_8783281_8783432 | 0.10 |
CAV3 |
caveolin 3 |
7804 |
0.14 |
| chr22_20008729_20009650 | 0.10 |
TANGO2 |
transport and golgi organization 2 homolog (Drosophila) |
511 |
0.64 |
| chr5_162992519_162993506 | 0.10 |
MAT2B |
methionine adenosyltransferase II, beta |
60392 |
0.11 |
| chr1_27183643_27183794 | 0.09 |
SFN |
stratifin |
5915 |
0.11 |
| chr11_73093533_73093684 | 0.09 |
RELT |
RELT tumor necrosis factor receptor |
5895 |
0.15 |
| chr8_142083713_142083864 | 0.09 |
DENND3 |
DENN/MADD domain containing 3 |
43589 |
0.14 |
| chr8_142141679_142142152 | 0.09 |
RP11-809O17.1 |
|
1855 |
0.33 |
| chr5_172659939_172660230 | 0.09 |
NKX2-5 |
NK2 homeobox 5 |
2125 |
0.3 |
| chr1_27720013_27720164 | 0.09 |
GPR3 |
G protein-coupled receptor 3 |
940 |
0.45 |
| chr1_109727416_109727567 | 0.09 |
ENSG00000238310 |
. |
22979 |
0.13 |
| chr11_3876946_3878257 | 0.08 |
STIM1 |
stromal interaction molecule 1 |
189 |
0.59 |
| chr5_140802398_140802934 | 0.08 |
PCDHGA11 |
protocadherin gamma subfamily A, 11 |
1871 |
0.13 |
| chr2_71126740_71126891 | 0.08 |
VAX2 |
ventral anterior homeobox 2 |
905 |
0.46 |
| chr7_45614270_45614575 | 0.08 |
ADCY1 |
adenylate cyclase 1 (brain) |
301 |
0.94 |
| chr17_4837485_4837705 | 0.08 |
GP1BA |
glycoprotein Ib (platelet), alpha polypeptide |
2003 |
0.12 |
| chr6_17101754_17102792 | 0.08 |
STMND1 |
stathmin domain containing 1 |
216 |
0.97 |
| chr5_176921947_176922464 | 0.08 |
RP11-1334A24.6 |
|
209 |
0.86 |
| chr19_574628_574849 | 0.08 |
BSG |
basigin (Ok blood group) |
309 |
0.77 |
| chr17_45730071_45730411 | 0.07 |
KPNB1 |
karyopherin (importin) beta 1 |
1679 |
0.27 |
| chr22_19566633_19566784 | 0.07 |
CLDN5 |
claudin 5 |
51640 |
0.11 |
| chr2_176969688_176969839 | 0.07 |
HOXD11 |
homeobox D11 |
2251 |
0.13 |
| chr22_17601345_17601496 | 0.07 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
723 |
0.48 |
| chr13_44360118_44360459 | 0.07 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
756 |
0.77 |
| chr16_69345315_69346083 | 0.07 |
VPS4A |
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
440 |
0.7 |
| chr22_50356186_50356903 | 0.07 |
PIM3 |
pim-3 oncogene |
2383 |
0.28 |
| chr7_155251163_155251329 | 0.07 |
EN2 |
engrailed homeobox 2 |
422 |
0.85 |
| chr16_73024055_73024442 | 0.07 |
ENSG00000221799 |
. |
5492 |
0.24 |
| chr22_50683947_50684307 | 0.07 |
TUBGCP6 |
tubulin, gamma complex associated protein 6 |
746 |
0.42 |
| chr11_125773182_125774068 | 0.07 |
PUS3 |
pseudouridylate synthase 3 |
509 |
0.49 |
| chr17_78233745_78234253 | 0.07 |
RNF213 |
ring finger protein 213 |
666 |
0.65 |
| chr15_80244332_80244483 | 0.06 |
BCL2A1 |
BCL2-related protein A1 |
19104 |
0.14 |
| chr12_30353916_30354159 | 0.06 |
ENSG00000251781 |
. |
105420 |
0.08 |
| chr14_51561527_51561763 | 0.06 |
TRIM9 |
tripartite motif containing 9 |
149 |
0.96 |
| chr3_157155512_157155783 | 0.06 |
PTX3 |
pentraxin 3, long |
1069 |
0.63 |
| chr19_8553014_8553200 | 0.06 |
HNRNPM |
heterogeneous nuclear ribonucleoprotein M |
2065 |
0.21 |
| chr4_23890798_23891066 | 0.06 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
726 |
0.8 |
| chr2_36877136_36877393 | 0.06 |
FEZ2 |
fasciculation and elongation protein zeta 2 (zygin II) |
4034 |
0.28 |
| chr1_201430589_201431038 | 0.06 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
7499 |
0.16 |
| chr7_27275476_27275737 | 0.05 |
EVX1 |
even-skipped homeobox 1 |
6558 |
0.1 |
| chr19_41168930_41169840 | 0.05 |
NUMBL |
numb homolog (Drosophila)-like |
19444 |
0.11 |
| chr15_69222833_69223294 | 0.05 |
NOX5 |
NADPH oxidase, EF-hand calcium binding domain 5 |
90 |
0.69 |
| chr5_16179066_16179217 | 0.05 |
MARCH11 |
membrane-associated ring finger (C3HC4) 11 |
743 |
0.56 |
| chr6_123317343_123317959 | 0.05 |
CLVS2 |
clavesin 2 |
183 |
0.97 |
| chr2_98962891_98963586 | 0.05 |
AC092675.4 |
|
220 |
0.79 |
| chr3_51989723_51989962 | 0.05 |
GPR62 |
G protein-coupled receptor 62 |
512 |
0.56 |
| chr1_186649228_186649406 | 0.05 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
242 |
0.96 |
| chr7_44529962_44530754 | 0.04 |
NUDCD3 |
NudC domain containing 3 |
121 |
0.96 |
| chr14_106331520_106331671 | 0.04 |
IGHJ1 |
immunoglobulin heavy joining 1 |
73 |
0.4 |
| chr16_2089839_2089990 | 0.04 |
SLC9A3R2 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
6642 |
0.06 |
| chr8_22224818_22225894 | 0.04 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
305 |
0.9 |
| chr17_73805350_73805708 | 0.04 |
UNK |
unkempt family zinc finger |
10291 |
0.09 |
| chr3_120626761_120627590 | 0.04 |
STXBP5L |
syntaxin binding protein 5-like |
14 |
0.99 |
| chr8_599967_600118 | 0.04 |
ERICH1 |
glutamate-rich 1 |
23615 |
0.22 |
| chr21_46902407_46902673 | 0.04 |
COL18A1 |
collagen, type XVIII, alpha 1 |
7649 |
0.21 |
| chr11_12028918_12029069 | 0.04 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
1137 |
0.63 |
| chr16_11295551_11295900 | 0.04 |
RMI2 |
RecQ mediated genome instability 2 |
47781 |
0.08 |
| chr6_26751971_26752122 | 0.04 |
ZNF322 |
zinc finger protein 322 |
92066 |
0.07 |
| chr12_71833793_71834525 | 0.04 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
344 |
0.86 |
| chr22_50709343_50709501 | 0.04 |
MAPK11 |
mitogen-activated protein kinase 11 |
600 |
0.52 |
| chr2_73518392_73518872 | 0.04 |
EGR4 |
early growth response 4 |
2197 |
0.23 |
| chr3_96531977_96533205 | 0.04 |
EPHA6 |
EPH receptor A6 |
834 |
0.76 |
| chr1_159906129_159906280 | 0.04 |
IGSF9 |
immunoglobulin superfamily, member 9 |
9182 |
0.1 |
| chr1_33116851_33117688 | 0.03 |
RBBP4 |
retinoblastoma binding protein 4 |
181 |
0.89 |
| chr4_102711575_102712644 | 0.03 |
BANK1 |
B-cell scaffold protein with ankyrin repeats 1 |
222 |
0.96 |
| chr7_27282525_27283078 | 0.03 |
EVX1 |
even-skipped homeobox 1 |
186 |
0.53 |
| chr2_200322083_200322803 | 0.03 |
SATB2-AS1 |
SATB2 antisense RNA 1 |
20 |
0.87 |
| chr15_79382979_79383271 | 0.03 |
RASGRF1 |
Ras protein-specific guanine nucleotide-releasing factor 1 |
10 |
0.98 |
| chr9_136738514_136738665 | 0.03 |
VAV2 |
vav 2 guanine nucleotide exchange factor |
118814 |
0.05 |
| chr19_16396913_16397119 | 0.03 |
CTD-2562J15.6 |
|
7370 |
0.17 |
| chr19_40926642_40927702 | 0.03 |
SERTAD1 |
SERTA domain containing 1 |
4760 |
0.11 |
| chr16_3043150_3043301 | 0.03 |
LA16c-380H5.2 |
|
11877 |
0.06 |
| chr2_149401643_149402183 | 0.03 |
EPC2 |
enhancer of polycomb homolog 2 (Drosophila) |
96 |
0.98 |
| chr10_70165992_70166978 | 0.03 |
RUFY2 |
RUN and FYVE domain containing 2 |
461 |
0.8 |
| chr1_206255126_206255277 | 0.03 |
ENSG00000252692 |
. |
5892 |
0.18 |
| chr17_40611024_40612223 | 0.03 |
ATP6V0A1 |
ATPase, H+ transporting, lysosomal V0 subunit a1 |
536 |
0.62 |
| chr16_1463725_1464471 | 0.03 |
UNKL |
unkempt family zinc finger-like |
594 |
0.51 |
| chr13_109924709_109925156 | 0.03 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
71101 |
0.13 |
| chr7_27190300_27190534 | 0.03 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
746 |
0.34 |
| chr18_74779548_74779839 | 0.03 |
MBP |
myelin basic protein |
37524 |
0.2 |
| chr11_116658098_116658624 | 0.03 |
ZNF259 |
zinc finger protein 259 |
334 |
0.79 |
| chr20_56286148_56286403 | 0.03 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
266 |
0.95 |
| chr17_32570563_32570714 | 0.03 |
CCL2 |
chemokine (C-C motif) ligand 2 |
11666 |
0.13 |
| chr8_9413349_9414633 | 0.03 |
RP11-375N15.2 |
|
39 |
0.92 |
| chr10_91294704_91295240 | 0.03 |
SLC16A12 |
solute carrier family 16, member 12 |
341 |
0.89 |
| chr2_42990357_42990508 | 0.03 |
OXER1 |
oxoeicosanoid (OXE) receptor 1 |
969 |
0.61 |
| chr19_17345728_17346858 | 0.03 |
OCEL1 |
occludin/ELL domain containing 1 |
7228 |
0.08 |
| chrX_118459285_118459436 | 0.03 |
RP5-1139I1.2 |
|
68198 |
0.08 |
| chr6_33558398_33558549 | 0.03 |
LINC00336 |
long intergenic non-protein coding RNA 336 |
2642 |
0.19 |
| chr15_75495247_75495814 | 0.03 |
C15orf39 |
chromosome 15 open reading frame 39 |
984 |
0.42 |
| chr7_132335536_132335698 | 0.03 |
AC009365.3 |
|
2064 |
0.31 |
| chr7_99159760_99160460 | 0.02 |
ZNF655 |
zinc finger protein 655 |
3625 |
0.14 |
| chr8_146156566_146156813 | 0.02 |
ZNF16 |
zinc finger protein 16 |
19292 |
0.14 |
| chr6_52977565_52977716 | 0.02 |
RP11-506E9.3 |
|
13304 |
0.12 |
| chr20_62517126_62517277 | 0.02 |
DNAJC5 |
DnaJ (Hsp40) homolog, subfamily C, member 5 |
9317 |
0.07 |
| chr9_35605722_35606427 | 0.02 |
TESK1 |
testis-specific kinase 1 |
707 |
0.45 |
| chr2_18479767_18479918 | 0.02 |
ENSG00000212455 |
. |
257843 |
0.02 |
| chr12_15881266_15881472 | 0.02 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
170 |
0.97 |
| chr16_22584462_22584656 | 0.02 |
RP11-368J21.3 |
|
26274 |
0.17 |
| chr5_178017459_178017882 | 0.02 |
COL23A1 |
collagen, type XXIII, alpha 1 |
114 |
0.97 |
| chr19_10229600_10230538 | 0.02 |
EIF3G |
eukaryotic translation initiation factor 3, subunit G |
471 |
0.6 |
| chr10_95326560_95326711 | 0.02 |
FFAR4 |
free fatty acid receptor 4 |
196 |
0.93 |
| chr1_17616214_17616365 | 0.02 |
ENSG00000266634 |
. |
11905 |
0.15 |
| chr16_89788705_89789477 | 0.02 |
ZNF276 |
zinc finger protein 276 |
1139 |
0.28 |
| chr4_55143470_55143900 | 0.02 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
33943 |
0.22 |
| chrX_118893537_118893849 | 0.02 |
SOWAHD |
sosondowah ankyrin repeat domain family member D |
1117 |
0.45 |
| chr18_657791_658473 | 0.02 |
C18orf56 |
chromosome 18 open reading frame 56 |
137 |
0.73 |
| chr19_33859506_33859657 | 0.02 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
4655 |
0.22 |
| chr19_708502_708753 | 0.02 |
PALM |
paralemmin |
326 |
0.8 |
| chr2_43232297_43232448 | 0.02 |
ENSG00000207087 |
. |
86260 |
0.1 |
| chr7_1552899_1553062 | 0.02 |
AC102953.6 |
|
3917 |
0.17 |
| chr9_81054275_81054473 | 0.02 |
PSAT1 |
phosphoserine aminotransferase 1 |
142315 |
0.05 |
| chr9_29213961_29214112 | 0.02 |
ENSG00000215939 |
. |
325083 |
0.01 |
| chr17_2308923_2309089 | 0.02 |
MNT |
MAX network transcriptional repressor |
4594 |
0.13 |
| chr20_44639888_44640039 | 0.02 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
2416 |
0.19 |
| chr2_8468645_8468796 | 0.02 |
AC011747.7 |
|
347176 |
0.01 |
| chr6_10882168_10882319 | 0.02 |
GCM2 |
glial cells missing homolog 2 (Drosophila) |
69 |
0.72 |
| chr15_66124802_66125877 | 0.02 |
ENSG00000252715 |
. |
10483 |
0.18 |
| chr11_64110647_64110798 | 0.02 |
CCDC88B |
coiled-coil domain containing 88B |
924 |
0.33 |
| chr8_145734508_145735192 | 0.02 |
MFSD3 |
major facilitator superfamily domain containing 3 |
393 |
0.6 |
| chr11_746807_747224 | 0.02 |
TALDO1 |
transaldolase 1 |
314 |
0.74 |
| chr1_204380536_204380863 | 0.01 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
220 |
0.93 |
| chr8_8820769_8821105 | 0.01 |
ENSG00000207244 |
. |
1852 |
0.35 |
| chr19_17649756_17650554 | 0.01 |
FAM129C |
family with sequence similarity 129, member C |
12042 |
0.1 |
| chr7_98028694_98029033 | 0.01 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
1517 |
0.48 |
| chr18_31803030_31803198 | 0.01 |
NOL4 |
nucleolar protein 4 |
55 |
0.68 |
| chr21_44013306_44013463 | 0.01 |
AP001625.6 |
|
31340 |
0.14 |
| chr2_230195712_230195863 | 0.01 |
PID1 |
phosphotyrosine interaction domain containing 1 |
59786 |
0.15 |
| chr17_73872840_73872991 | 0.01 |
TRIM47 |
tripartite motif containing 47 |
430 |
0.42 |
| chr9_124988280_124988557 | 0.01 |
LHX6 |
LIM homeobox 6 |
1447 |
0.33 |
| chr8_143809061_143809726 | 0.01 |
CTD-2292P10.2 |
|
285 |
0.66 |
| chr9_131038025_131038209 | 0.01 |
GOLGA2 |
golgin A2 |
102 |
0.73 |
| chr2_75072423_75073024 | 0.01 |
HK2 |
hexokinase 2 |
10426 |
0.25 |
| chr7_128337244_128337509 | 0.01 |
ENSG00000201041 |
. |
117 |
0.79 |
| chr19_34973467_34973843 | 0.01 |
WTIP |
Wilms tumor 1 interacting protein |
222 |
0.92 |
| chr10_47767707_47767858 | 0.01 |
ANXA8L2 |
annexin A8-like 2 |
20746 |
0.17 |
| chr20_61846294_61847306 | 0.01 |
YTHDF1 |
YTH domain family, member 1 |
691 |
0.57 |
| chr8_43046584_43046788 | 0.01 |
HGSNAT |
heparan-alpha-glucosaminide N-acetyltransferase |
11254 |
0.22 |
| chr4_113778484_113778635 | 0.01 |
RP11-119H12.6 |
|
24927 |
0.19 |
| chr5_179740873_179741196 | 0.01 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
17614 |
0.17 |
| chr22_37921660_37921818 | 0.01 |
CARD10 |
caspase recruitment domain family, member 10 |
6190 |
0.14 |
| chr1_149871196_149871937 | 0.01 |
BOLA1 |
bolA family member 1 |
387 |
0.67 |
| chr17_70511481_70511655 | 0.01 |
ENSG00000200783 |
. |
148723 |
0.04 |
| chr7_138720144_138720977 | 0.01 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
215 |
0.95 |
| chr11_18405572_18406024 | 0.01 |
ENSG00000264603 |
. |
3536 |
0.15 |
| chr6_158423816_158423967 | 0.01 |
SYNJ2-IT1 |
SYNJ2 intronic transcript 1 (non-protein coding) |
1752 |
0.37 |
| chr10_48276074_48276225 | 0.01 |
ANXA8 |
annexin A8 |
20796 |
0.13 |
| chr20_35484644_35484795 | 0.01 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
3557 |
0.21 |
| chr1_89536224_89536375 | 0.01 |
GBP1 |
guanylate binding protein 1, interferon-inducible |
5256 |
0.19 |
| chr19_46366604_46366815 | 0.01 |
SYMPK |
symplekin |
161 |
0.8 |
| chr19_13231168_13231373 | 0.01 |
NACC1 |
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
2119 |
0.16 |
| chr5_77840590_77840741 | 0.01 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
4309 |
0.35 |
| chr7_5820371_5821002 | 0.01 |
RNF216 |
ring finger protein 216 |
565 |
0.76 |
| chr2_129166329_129166512 | 0.01 |
ENSG00000238379 |
. |
36352 |
0.19 |
| chr3_75690482_75691208 | 0.01 |
ENSG00000221795 |
. |
10931 |
0.2 |
| chr3_37670033_37670331 | 0.01 |
ENSG00000239105 |
. |
55478 |
0.15 |
| chr8_82753589_82754405 | 0.01 |
SNX16 |
sorting nexin 16 |
196 |
0.96 |
| chr5_115151846_115152134 | 0.01 |
CDO1 |
cysteine dioxygenase type 1 |
661 |
0.69 |
| chr11_280225_280457 | 0.01 |
NLRP6 |
NLR family, pyrin domain containing 6 |
1771 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.2 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |