Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
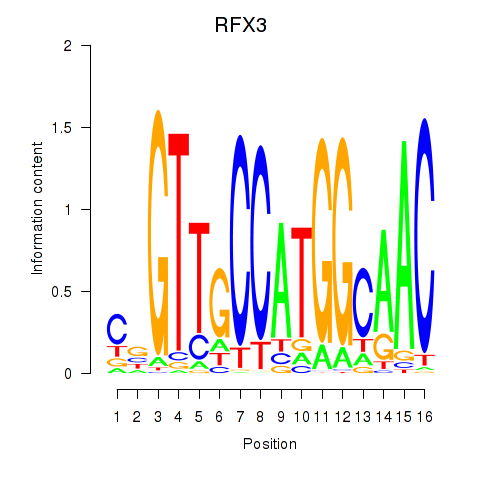
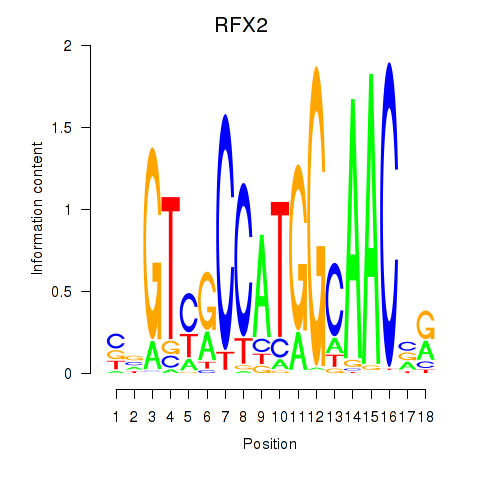
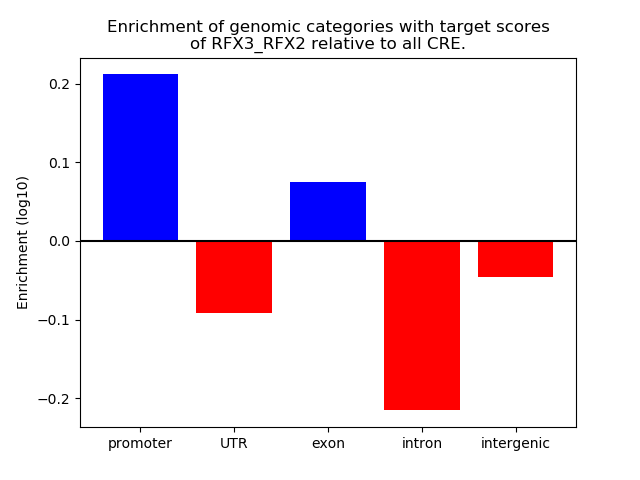
Results for RFX3_RFX2
Z-value: 1.68
Transcription factors associated with RFX3_RFX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX3
|
ENSG00000080298.11 | regulatory factor X3 |
|
RFX2
|
ENSG00000087903.8 | regulatory factor X2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_6060606_6060757 | RFX2 | 3387 | 0.184856 | -0.65 | 5.9e-02 | Click! |
| chr19_6109831_6110198 | RFX2 | 443 | 0.583370 | -0.62 | 7.3e-02 | Click! |
| chr19_6060334_6060560 | RFX2 | 3153 | 0.191533 | -0.62 | 7.3e-02 | Click! |
| chr19_6061582_6061733 | RFX2 | 4363 | 0.166615 | -0.58 | 1.0e-01 | Click! |
| chr19_6023295_6023446 | RFX2 | 15133 | 0.130433 | 0.57 | 1.1e-01 | Click! |
| chr9_3241640_3241791 | RFX3 | 15484 | 0.298043 | 0.67 | 4.7e-02 | Click! |
| chr9_3342385_3342536 | RFX3 | 4304 | 0.355989 | 0.50 | 1.7e-01 | Click! |
| chr9_3525532_3525838 | RFX3 | 298 | 0.941962 | 0.49 | 1.8e-01 | Click! |
| chr9_3398052_3398553 | RFX3 | 2706 | 0.398440 | -0.45 | 2.2e-01 | Click! |
| chr9_3507919_3508070 | RFX3 | 17989 | 0.248199 | -0.42 | 2.5e-01 | Click! |
Activity of the RFX3_RFX2 motif across conditions
Conditions sorted by the z-value of the RFX3_RFX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
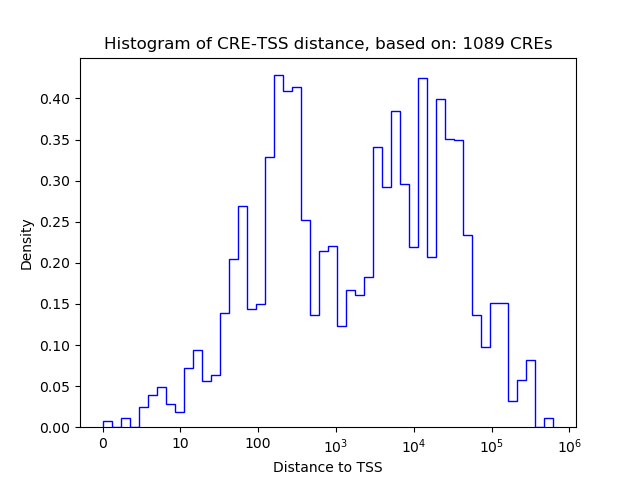
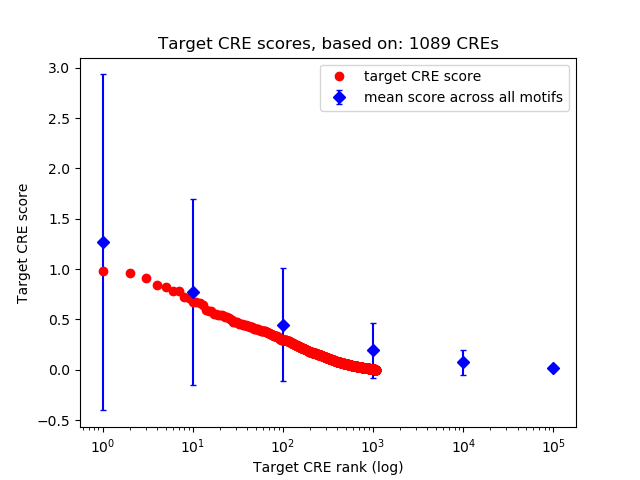
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr20_36888706_36889089 | 0.98 |
KIAA1755 |
KIAA1755 |
277 |
0.53 |
| chr5_16713576_16713796 | 0.96 |
MYO10 |
myosin X |
24810 |
0.21 |
| chr17_60885608_60885921 | 0.91 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
59 |
0.98 |
| chr17_45785766_45785998 | 0.85 |
TBKBP1 |
TBK1 binding protein 1 |
13252 |
0.13 |
| chr11_8040357_8041136 | 0.82 |
TUB |
tubby bipartite transcription factor |
45 |
0.96 |
| chr12_80794329_80794553 | 0.78 |
PTPRQ |
protein tyrosine phosphatase, receptor type, Q |
5333 |
0.26 |
| chr14_23355781_23356199 | 0.78 |
REM2 |
RAS (RAD and GEM)-like GTP binding 2 |
3559 |
0.1 |
| chr7_134468265_134468416 | 0.72 |
CALD1 |
caldesmon 1 |
3911 |
0.34 |
| chr19_14048794_14049313 | 0.71 |
PODNL1 |
podocan-like 1 |
144 |
0.92 |
| chr10_80008606_80008848 | 0.67 |
ENSG00000201393 |
. |
118537 |
0.06 |
| chr1_44031509_44031896 | 0.67 |
PTPRF |
protein tyrosine phosphatase, receptor type, F |
20955 |
0.18 |
| chr6_112823116_112823311 | 0.67 |
ENSG00000239095 |
. |
25285 |
0.2 |
| chr16_89990965_89991193 | 0.65 |
TUBB3 |
Tubulin beta-3 chain |
1305 |
0.26 |
| chr5_176873858_176874453 | 0.59 |
PRR7 |
proline rich 7 (synaptic) |
327 |
0.6 |
| chr14_90989296_90989447 | 0.58 |
ENSG00000252748 |
. |
42114 |
0.16 |
| chr14_69052548_69052699 | 0.58 |
CTD-2325P2.4 |
|
42539 |
0.18 |
| chr9_86886547_86886817 | 0.56 |
RP11-380F14.2 |
|
6452 |
0.29 |
| chr6_161664088_161664497 | 0.55 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
30759 |
0.25 |
| chrX_53078377_53078712 | 0.55 |
GPR173 |
G protein-coupled receptor 173 |
79 |
0.97 |
| chr17_65524931_65525141 | 0.54 |
CTD-2653B5.1 |
|
4439 |
0.23 |
| chr12_48576703_48577574 | 0.54 |
C12orf68 |
chromosome 12 open reading frame 68 |
228 |
0.88 |
| chrX_10584505_10584656 | 0.53 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
3879 |
0.32 |
| chr5_174486834_174487088 | 0.53 |
ENSG00000266890 |
. |
308224 |
0.01 |
| chr20_17816826_17817060 | 0.52 |
ENSG00000221220 |
. |
6284 |
0.24 |
| chr9_94579821_94580301 | 0.51 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
131101 |
0.05 |
| chr2_8818889_8820001 | 0.50 |
AC011747.7 |
|
259 |
0.7 |
| chr4_140823783_140824037 | 0.50 |
MAML3 |
mastermind-like 3 (Drosophila) |
11789 |
0.28 |
| chr7_73448257_73448408 | 0.48 |
ELN |
elastin |
5814 |
0.21 |
| chr2_5979840_5979991 | 0.48 |
DKFZP761K2322 |
HCG1990367; Putative uncharacterized protein DKFZp761K2322; Putative uncharacterized protein FLJ30594; Uncharacterized protein; cDNA FLJ30594 fis, clone BRAWH2008903 |
131797 |
0.05 |
| chr18_21562271_21562511 | 0.47 |
TTC39C |
tetratricopeptide repeat domain 39C |
10346 |
0.18 |
| chr8_74282720_74282947 | 0.47 |
RP11-434I12.2 |
|
14137 |
0.25 |
| chr2_187713432_187714439 | 0.46 |
ZSWIM2 |
zinc finger, SWIM-type containing 2 |
38 |
0.99 |
| chr1_84630171_84630563 | 0.46 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
3 |
0.99 |
| chr4_140737608_140737759 | 0.46 |
ENSG00000252233 |
. |
22096 |
0.23 |
| chr15_65341843_65342117 | 0.45 |
SLC51B |
solute carrier family 51, beta subunit |
4272 |
0.15 |
| chr17_64187141_64188685 | 0.45 |
CEP112 |
centrosomal protein 112kDa |
60 |
0.98 |
| chr11_101918104_101918467 | 0.45 |
C11orf70 |
chromosome 11 open reading frame 70 |
96 |
0.96 |
| chr22_20143038_20143223 | 0.45 |
AC006547.14 |
uncharacterized protein LOC388849 |
4731 |
0.11 |
| chr4_96102666_96102817 | 0.44 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
77235 |
0.13 |
| chr17_19314379_19314710 | 0.44 |
RNF112 |
ring finger protein 112 |
37 |
0.95 |
| chr17_76950200_76950351 | 0.44 |
LGALS3BP |
lectin, galactoside-binding, soluble, 3 binding protein |
23130 |
0.12 |
| chr1_205283210_205283468 | 0.43 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
7544 |
0.17 |
| chr5_139418008_139418169 | 0.43 |
NRG2 |
neuregulin 2 |
4566 |
0.2 |
| chr3_49055259_49056395 | 0.43 |
DALRD3 |
DALR anticodon binding domain containing 3 |
170 |
0.84 |
| chr1_85928394_85928655 | 0.43 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
1743 |
0.41 |
| chr2_175870043_175870550 | 0.43 |
CHN1 |
chimerin 1 |
199 |
0.93 |
| chr4_170189368_170189538 | 0.42 |
SH3RF1 |
SH3 domain containing ring finger 1 |
1655 |
0.5 |
| chr15_57109701_57109852 | 0.41 |
ZNF280D |
zinc finger protein 280D |
83492 |
0.1 |
| chr1_111214730_111214881 | 0.41 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
2850 |
0.25 |
| chr10_43818085_43818364 | 0.41 |
ENSG00000221468 |
. |
19010 |
0.18 |
| chr10_26829653_26829804 | 0.40 |
ENSG00000199733 |
. |
31210 |
0.22 |
| chr1_54940257_54940662 | 0.40 |
ENSG00000265404 |
. |
29882 |
0.15 |
| chr12_122110363_122110677 | 0.40 |
MORN3 |
MORN repeat containing 3 |
2960 |
0.25 |
| chr11_132934198_132934477 | 0.40 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
120674 |
0.06 |
| chr19_5293472_5293908 | 0.39 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
427 |
0.88 |
| chr8_42010434_42011226 | 0.39 |
AP3M2 |
adaptor-related protein complex 3, mu 2 subunit |
168 |
0.83 |
| chr5_61389807_61390019 | 0.39 |
ENSG00000251983 |
. |
52579 |
0.18 |
| chr8_120428169_120429285 | 0.39 |
NOV |
nephroblastoma overexpressed |
181 |
0.96 |
| chr11_132813791_132813979 | 0.39 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
222 |
0.96 |
| chr11_45921095_45921634 | 0.39 |
MAPK8IP1 |
mitogen-activated protein kinase 8 interacting protein 1 |
3263 |
0.15 |
| chr10_6316976_6317295 | 0.39 |
ENSG00000238366 |
. |
29341 |
0.16 |
| chr7_158766053_158766279 | 0.38 |
ENSG00000231419 |
. |
34946 |
0.21 |
| chrX_47175743_47175914 | 0.38 |
ENSG00000266158 |
. |
35366 |
0.1 |
| chr10_103599730_103600193 | 0.38 |
KCNIP2 |
Kv channel interacting protein 2 |
332 |
0.86 |
| chr3_196729783_196730213 | 0.38 |
MFI2-AS1 |
MFI2 antisense RNA 1 |
5 |
0.97 |
| chr22_41110494_41110716 | 0.37 |
MCHR1 |
melanin-concentrating hormone receptor 1 |
35328 |
0.15 |
| chr9_19493377_19493724 | 0.37 |
RP11-363E7.4 |
|
40343 |
0.16 |
| chr19_10678474_10679697 | 0.37 |
CDKN2D |
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
569 |
0.54 |
| chr14_24838154_24838623 | 0.37 |
NFATC4 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 |
15 |
0.95 |
| chr1_155043063_155043480 | 0.37 |
EFNA4 |
ephrin-A4 |
7034 |
0.08 |
| chr15_84748384_84748959 | 0.36 |
EFTUD1P1 |
elongation factor Tu GTP binding domain containing 1 pseudogene 1 |
249 |
0.94 |
| chr20_291107_291258 | 0.36 |
ZCCHC3 |
zinc finger, CCHC domain containing 3 |
13445 |
0.11 |
| chr7_157361164_157361701 | 0.36 |
ENSG00000207960 |
. |
5682 |
0.22 |
| chr1_204034501_204034712 | 0.35 |
SOX13 |
SRY (sex determining region Y)-box 13 |
8126 |
0.2 |
| chr19_46919550_46919940 | 0.35 |
CCDC8 |
coiled-coil domain containing 8 |
2904 |
0.19 |
| chr1_242097620_242097936 | 0.34 |
EXO1 |
exonuclease 1 |
55436 |
0.13 |
| chr16_72965340_72965833 | 0.34 |
ENSG00000221799 |
. |
53170 |
0.13 |
| chr5_111890138_111890295 | 0.34 |
EPB41L4A-AS2 |
EPB41L4A antisense RNA 2 (head to head) |
134936 |
0.05 |
| chr6_132268739_132268890 | 0.34 |
RP11-69I8.3 |
|
3272 |
0.26 |
| chr22_43539145_43539583 | 0.34 |
MCAT |
malonyl CoA:ACP acyltransferase (mitochondrial) |
6 |
0.97 |
| chr12_56131718_56132374 | 0.34 |
GDF11 |
growth differentiation factor 11 |
5018 |
0.09 |
| chr1_203445281_203445432 | 0.34 |
PRELP |
proline/arginine-rich end leucine-rich repeat protein |
400 |
0.87 |
| chr1_33546320_33546691 | 0.33 |
ADC |
arginine decarboxylase |
209 |
0.94 |
| chrX_102809591_102809942 | 0.33 |
TCEAL4 |
transcription elongation factor A (SII)-like 4 |
21393 |
0.13 |
| chr8_145027093_145027557 | 0.33 |
PLEC |
plectin |
763 |
0.48 |
| chr5_122181350_122181750 | 0.33 |
SNX24 |
sorting nexin 24 |
229 |
0.93 |
| chr1_244175541_244175807 | 0.33 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
38911 |
0.17 |
| chr1_179050699_179051584 | 0.33 |
TOR3A |
torsin family 3, member A |
64 |
0.98 |
| chr10_79006464_79006615 | 0.32 |
RP11-328K22.1 |
|
67160 |
0.12 |
| chr7_1181998_1182149 | 0.32 |
C7orf50 |
chromosome 7 open reading frame 50 |
4177 |
0.16 |
| chr4_186853773_186853924 | 0.31 |
ENSG00000239034 |
. |
13389 |
0.21 |
| chr8_102092961_102093560 | 0.31 |
ENSG00000202360 |
. |
56927 |
0.13 |
| chr6_3179788_3179988 | 0.31 |
TUBB2A |
tubulin, beta 2A class IIa |
22128 |
0.13 |
| chr17_37388032_37388399 | 0.30 |
STAC2 |
SH3 and cysteine rich domain 2 |
6090 |
0.15 |
| chr4_186125321_186125752 | 0.30 |
SNX25 |
sorting nexin 25 |
145 |
0.75 |
| chr18_43206329_43206480 | 0.30 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
11638 |
0.2 |
| chr2_242606070_242606221 | 0.30 |
ATG4B |
autophagy related 4B, cysteine peptidase |
134 |
0.93 |
| chr17_46089516_46089790 | 0.30 |
RP11-6N17.10 |
|
16091 |
0.08 |
| chr6_164506356_164506803 | 0.30 |
ENSG00000266128 |
. |
244986 |
0.02 |
| chr12_94580654_94580885 | 0.30 |
RP11-74K11.2 |
|
949 |
0.61 |
| chr3_158288685_158288908 | 0.30 |
RP11-538P18.2 |
|
59 |
0.64 |
| chr1_173638556_173639045 | 0.30 |
ANKRD45 |
ankyrin repeat domain 45 |
201 |
0.93 |
| chr1_15573540_15574233 | 0.30 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
110 |
0.97 |
| chr18_44526809_44527253 | 0.30 |
KATNAL2 |
katanin p60 subunit A-like 2 |
213 |
0.91 |
| chr8_144302159_144302531 | 0.29 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
7277 |
0.14 |
| chr15_42066837_42067403 | 0.29 |
MAPKBP1 |
mitogen-activated protein kinase binding protein 1 |
220 |
0.9 |
| chr4_87280706_87281702 | 0.29 |
MAPK10 |
mitogen-activated protein kinase 10 |
12 |
0.98 |
| chr7_136795005_136795156 | 0.29 |
hsa-mir-490 |
hsa-mir-490 |
53937 |
0.17 |
| chr12_109548691_109549386 | 0.29 |
RP11-968O1.5 |
|
15 |
0.96 |
| chr15_92237487_92237638 | 0.29 |
ENSG00000238981 |
. |
141047 |
0.05 |
| chr13_24554384_24554726 | 0.29 |
SPATA13 |
spermatogenesis associated 13 |
611 |
0.77 |
| chr11_8832007_8832297 | 0.29 |
ST5 |
suppression of tumorigenicity 5 |
44 |
0.97 |
| chr21_44061431_44061649 | 0.29 |
AP001626.2 |
|
9814 |
0.18 |
| chr19_38705050_38705201 | 0.29 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
8688 |
0.1 |
| chr22_37448126_37448435 | 0.29 |
KCTD17 |
potassium channel tetramerization domain containing 17 |
70 |
0.94 |
| chr12_7037386_7038830 | 0.28 |
ATN1 |
atrophin 1 |
632 |
0.41 |
| chr9_82278752_82279150 | 0.28 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
11443 |
0.33 |
| chr19_47249862_47250221 | 0.28 |
STRN4 |
striatin, calmodulin binding protein 4 |
97 |
0.53 |
| chr6_32119881_32120041 | 0.28 |
PRRT1 |
proline-rich transmembrane protein 1 |
82 |
0.9 |
| chr2_220083227_220083846 | 0.28 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
136 |
0.88 |
| chr19_52193570_52193969 | 0.27 |
ENSG00000207550 |
. |
2096 |
0.15 |
| chr17_39560147_39560298 | 0.27 |
AC003958.2 |
|
1547 |
0.2 |
| chr16_57662077_57662305 | 0.27 |
GPR56 |
G protein-coupled receptor 56 |
21 |
0.97 |
| chr6_43612507_43612686 | 0.27 |
RSPH9 |
radial spoke head 9 homolog (Chlamydomonas) |
187 |
0.91 |
| chr1_179783744_179784035 | 0.27 |
RP11-12M5.3 |
|
2670 |
0.26 |
| chrX_70365452_70365975 | 0.27 |
NLGN3 |
neuroligin 3 |
1001 |
0.39 |
| chr3_156874639_156874950 | 0.27 |
CCNL1 |
cyclin L1 |
3208 |
0.18 |
| chr3_45068445_45068596 | 0.26 |
CLEC3B |
C-type lectin domain family 3, member B |
845 |
0.58 |
| chr7_50850197_50851001 | 0.26 |
GRB10 |
growth factor receptor-bound protein 10 |
335 |
0.93 |
| chr8_22860264_22860415 | 0.26 |
PEBP4 |
phosphatidylethanolamine-binding protein 4 |
2826 |
0.17 |
| chr7_129781517_129782004 | 0.26 |
RP11-775D22.2 |
|
182 |
0.94 |
| chr16_2892196_2892619 | 0.26 |
ZG16B |
zymogen granule protein 16B |
12038 |
0.08 |
| chr8_142141364_142141568 | 0.25 |
RP11-809O17.1 |
|
1406 |
0.4 |
| chr6_28979234_28979409 | 0.25 |
ZNF311 |
zinc finger protein 311 |
6284 |
0.13 |
| chr1_148532242_148532716 | 0.25 |
NBPF15 |
neuroblastoma breakpoint family, member 15 |
28364 |
0.22 |
| chr1_198650996_198651147 | 0.25 |
RP11-553K8.5 |
|
14881 |
0.25 |
| chr12_48690908_48691108 | 0.25 |
H1FNT |
H1 histone family, member N, testis-specific |
31755 |
0.12 |
| chr3_38080498_38081224 | 0.25 |
DLEC1 |
deleted in lung and esophageal cancer 1 |
165 |
0.94 |
| chr19_48948815_48949184 | 0.24 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
31 |
0.95 |
| chr16_67564286_67564635 | 0.24 |
FAM65A |
family with sequence similarity 65, member A |
766 |
0.44 |
| chr19_47353649_47354096 | 0.24 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
151 |
0.95 |
| chr1_15852991_15853242 | 0.24 |
DNAJC16 |
DnaJ (Hsp40) homolog, subfamily C, member 16 |
192 |
0.89 |
| chr14_75777417_75777662 | 0.24 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
30643 |
0.14 |
| chr17_70338672_70338823 | 0.24 |
SOX9 |
SRY (sex determining region Y)-box 9 |
221586 |
0.02 |
| chr2_174855935_174856086 | 0.24 |
SP3 |
Sp3 transcription factor |
25580 |
0.26 |
| chr5_140893341_140893614 | 0.24 |
DIAPH1 |
diaphanous-related formin 1 |
12237 |
0.09 |
| chr12_58146355_58146506 | 0.24 |
CDK4 |
cyclin-dependent kinase 4 |
126 |
0.88 |
| chr17_41176429_41177125 | 0.23 |
VAT1 |
vesicle amine transport 1 |
362 |
0.51 |
| chr15_40762663_40763059 | 0.23 |
CHST14 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
299 |
0.8 |
| chr16_4049606_4049757 | 0.23 |
ADCY9 |
adenylate cyclase 9 |
10680 |
0.18 |
| chr19_19729670_19729882 | 0.23 |
PBX4 |
pre-B-cell leukemia homeobox 4 |
51 |
0.96 |
| chr12_2921492_2921671 | 0.23 |
RP4-816N1.6 |
|
34 |
0.71 |
| chr8_6407769_6407977 | 0.23 |
ANGPT2 |
angiopoietin 2 |
12692 |
0.26 |
| chr3_57529834_57530279 | 0.22 |
DNAH12 |
dynein, axonemal, heavy chain 12 |
12 |
0.96 |
| chr3_125321989_125322196 | 0.22 |
OSBPL11 |
oxysterol binding protein-like 11 |
8158 |
0.22 |
| chr22_25506820_25506976 | 0.22 |
CTA-221G9.11 |
|
1761 |
0.41 |
| chr1_144573049_144573284 | 0.22 |
ENSG00000201699 |
. |
39128 |
0.14 |
| chr16_30568785_30569715 | 0.22 |
ZNF764 |
zinc finger protein 764 |
334 |
0.43 |
| chr6_35181325_35181573 | 0.22 |
SCUBE3 |
signal peptide, CUB domain, EGF-like 3 |
741 |
0.69 |
| chr2_132285357_132286129 | 0.22 |
CCDC74A |
coiled-coil domain containing 74A |
290 |
0.91 |
| chr10_120101830_120102206 | 0.22 |
FAM204A |
family with sequence similarity 204, member A |
186 |
0.97 |
| chr7_48075055_48075727 | 0.22 |
C7orf57 |
chromosome 7 open reading frame 57 |
274 |
0.91 |
| chr5_61708037_61708393 | 0.22 |
IPO11 |
importin 11 |
273 |
0.91 |
| chr8_22462053_22462924 | 0.22 |
CCAR2 |
cell cycle and apoptosis regulator 2 |
63 |
0.94 |
| chr1_201600215_201600366 | 0.22 |
NAV1 |
neuron navigator 1 |
7689 |
0.16 |
| chr17_7117806_7118379 | 0.22 |
DLG4 |
discs, large homolog 4 (Drosophila) |
861 |
0.31 |
| chr9_34620609_34620827 | 0.21 |
DCTN3 |
dynactin 3 (p22) |
203 |
0.86 |
| chr14_57046308_57046738 | 0.21 |
TMEM260 |
transmembrane protein 260 |
12 |
0.99 |
| chr14_59628263_59628414 | 0.21 |
DAAM1 |
dishevelled associated activator of morphogenesis 1 |
27060 |
0.26 |
| chr7_5184079_5184242 | 0.21 |
ZNF890P |
zinc finger protein 890, pseudogene |
16734 |
0.17 |
| chr18_380848_380999 | 0.21 |
RP11-720L2.2 |
|
43493 |
0.16 |
| chr21_39286916_39287067 | 0.21 |
KCNJ6 |
potassium inwardly-rectifying channel, subfamily J, member 6 |
1434 |
0.51 |
| chr17_60947671_60947822 | 0.21 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
62041 |
0.12 |
| chr2_130901985_130902795 | 0.21 |
CCDC74B |
coiled-coil domain containing 74B |
177 |
0.9 |
| chr2_3200081_3200232 | 0.20 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
105080 |
0.07 |
| chr21_33952233_33952384 | 0.20 |
TCP10L |
t-complex 10-like |
5535 |
0.15 |
| chr12_51818248_51818602 | 0.20 |
RP11-607P23.1 |
|
30 |
0.62 |
| chr8_145024226_145024570 | 0.20 |
PLEC |
plectin |
646 |
0.55 |
| chr17_76320400_76320721 | 0.20 |
SOCS3 |
suppressor of cytokine signaling 3 |
35595 |
0.11 |
| chr1_183440636_183441395 | 0.20 |
SMG7 |
SMG7 nonsense mediated mRNA decay factor |
491 |
0.83 |
| chrX_71351813_71352291 | 0.20 |
RGAG4 |
retrotransposon gag domain containing 4 |
294 |
0.84 |
| chr7_1576476_1576960 | 0.20 |
MAFK |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog K |
1626 |
0.3 |
| chr5_148521385_148522083 | 0.20 |
ABLIM3 |
actin binding LIM protein family, member 3 |
124 |
0.96 |
| chr1_172352648_172352805 | 0.20 |
DNM3 |
dynamin 3 |
4568 |
0.19 |
| chr2_178482857_178483675 | 0.20 |
TTC30A |
tetratricopeptide repeat domain 30A |
428 |
0.84 |
| chr11_47600251_47600536 | 0.20 |
KBTBD4 |
kelch repeat and BTB (POZ) domain containing 4 |
52 |
0.64 |
| chr3_138047908_138048749 | 0.19 |
NME9 |
NME/NM23 family member 9 |
123 |
0.97 |
| chr12_106736611_106736853 | 0.19 |
POLR3B |
polymerase (RNA) III (DNA directed) polypeptide B |
14704 |
0.19 |
| chr14_59931736_59932162 | 0.19 |
GPR135 |
G protein-coupled receptor 135 |
111 |
0.97 |
| chr9_35828652_35829114 | 0.19 |
FAM221B |
family with sequence similarity 221, member B |
154 |
0.7 |
| chr8_22084641_22084931 | 0.19 |
PHYHIP |
phytanoyl-CoA 2-hydroxylase interacting protein |
420 |
0.76 |
| chr10_31891715_31891977 | 0.19 |
ENSG00000222412 |
. |
153218 |
0.04 |
| chr19_56826038_56826226 | 0.18 |
ZSCAN5A |
zinc finger and SCAN domain containing 5A |
25 |
0.51 |
| chr9_140024279_140024727 | 0.18 |
ENSG00000238824 |
. |
6443 |
0.06 |
| chr6_139644906_139645379 | 0.18 |
TXLNB |
taxilin beta |
31866 |
0.17 |
| chr6_30640170_30640817 | 0.18 |
DHX16 |
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
272 |
0.78 |
| chr19_37803327_37803721 | 0.18 |
HKR1 |
HKR1, GLI-Kruppel zinc finger family member |
215 |
0.93 |
| chr21_44061692_44061930 | 0.18 |
AP001626.2 |
|
9543 |
0.18 |
| chr2_3286362_3286843 | 0.18 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
18634 |
0.24 |
| chr18_47833726_47834243 | 0.18 |
ENSG00000251997 |
. |
11388 |
0.15 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.1 | 0.3 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.3 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.4 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.1 | 0.5 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.8 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.2 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.5 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.3 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.0 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.0 | GO:0035583 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0000080 | mitotic G1 phase(GO:0000080) |
| 0.0 | 0.0 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.1 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.3 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.3 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 0.4 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0042171 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0032142 | single guanine insertion binding(GO:0032142) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0046978 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME GABA RECEPTOR ACTIVATION | Genes involved in GABA receptor activation |