Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
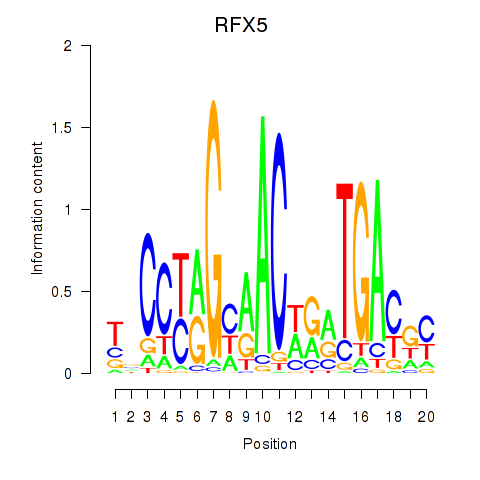
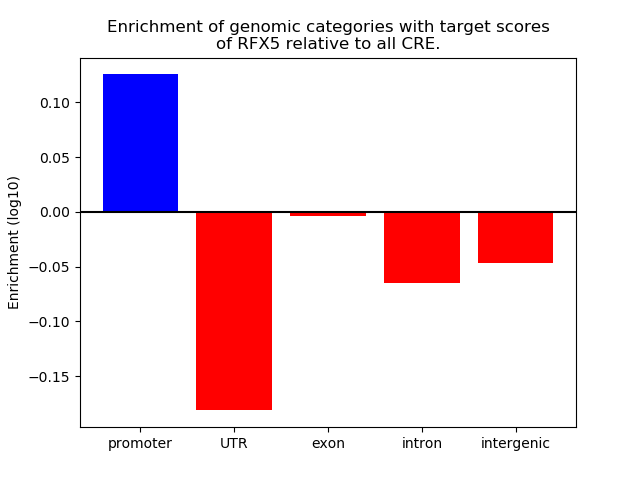
Results for RFX5
Z-value: 0.53
Transcription factors associated with RFX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RFX5
|
ENSG00000143390.13 | regulatory factor X5 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_151318667_151318818 | RFX5 | 340 | 0.637033 | 0.32 | 4.1e-01 | Click! |
| chr1_151318943_151319712 | RFX5 | 21 | 0.592683 | 0.15 | 6.9e-01 | Click! |
| chr1_151319989_151320443 | RFX5 | 383 | 0.611252 | -0.12 | 7.6e-01 | Click! |
Activity of the RFX5 motif across conditions
Conditions sorted by the z-value of the RFX5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
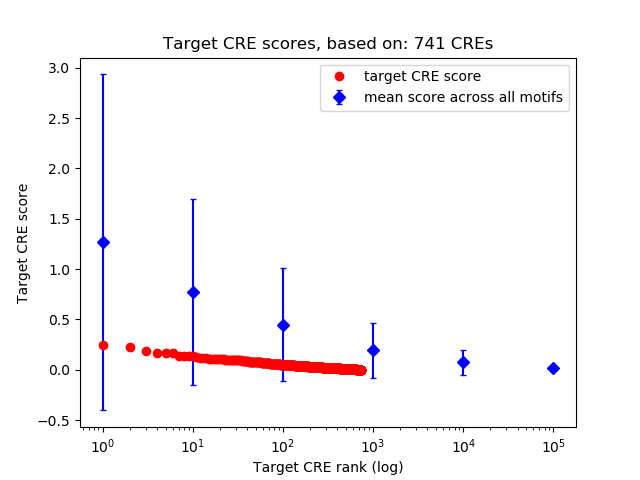
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_157893196_157893594 | 0.24 |
PDGFC |
platelet derived growth factor C |
849 |
0.66 |
| chr3_49841438_49841709 | 0.23 |
FAM212A |
family with sequence similarity 212, member A |
886 |
0.35 |
| chr20_45980431_45980900 | 0.19 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
44 |
0.91 |
| chr6_45404317_45404604 | 0.17 |
RUNX2 |
runt-related transcription factor 2 |
14238 |
0.21 |
| chr1_153958983_153959396 | 0.17 |
RAB13 |
RAB13, member RAS oncogene family |
361 |
0.72 |
| chr2_165478229_165478434 | 0.16 |
GRB14 |
growth factor receptor-bound protein 14 |
27 |
0.99 |
| chr17_67605378_67605727 | 0.14 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
106982 |
0.08 |
| chr7_101765454_101765681 | 0.14 |
ENSG00000252824 |
. |
71877 |
0.09 |
| chr2_66782749_66783275 | 0.14 |
MEIS1 |
Meis homeobox 1 |
46953 |
0.17 |
| chr12_48170565_48170736 | 0.13 |
SLC48A1 |
solute carrier family 48 (heme transporter), member 1 |
3659 |
0.16 |
| chr18_77492818_77492969 | 0.13 |
RP11-567M16.5 |
|
29242 |
0.2 |
| chr22_37938188_37938339 | 0.12 |
RP5-1177I5.3 |
|
7770 |
0.13 |
| chr13_95619763_95620412 | 0.12 |
ENSG00000252335 |
. |
51402 |
0.18 |
| chr14_76072100_76072251 | 0.11 |
FLVCR2 |
feline leukemia virus subgroup C cellular receptor family, member 2 |
17 |
0.97 |
| chr20_35483169_35483320 | 0.11 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
5032 |
0.18 |
| chr5_177018982_177019137 | 0.11 |
TMED9 |
transmembrane emp24 protein transport domain containing 9 |
100 |
0.95 |
| chr20_45946936_45948261 | 0.10 |
AL031666.2 |
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
352 |
0.82 |
| chr2_217702830_217702981 | 0.10 |
AC007557.4 |
|
16950 |
0.12 |
| chr13_73633121_73633496 | 0.10 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
378 |
0.89 |
| chr19_49199645_49199859 | 0.10 |
FUT2 |
fucosyltransferase 2 (secretor status included) |
520 |
0.59 |
| chr4_140579390_140579541 | 0.10 |
MGST2 |
microsomal glutathione S-transferase 2 |
7457 |
0.21 |
| chr20_35152594_35152745 | 0.10 |
RP5-977B1.7 |
|
1864 |
0.29 |
| chr13_100160443_100160594 | 0.10 |
TM9SF2 |
transmembrane 9 superfamily member 2 |
6847 |
0.17 |
| chr15_90283484_90283688 | 0.10 |
MESP1 |
mesoderm posterior 1 homolog (mouse) |
10955 |
0.11 |
| chr8_41907411_41908748 | 0.10 |
KAT6A |
K(lysine) acetyltransferase 6A |
1426 |
0.43 |
| chr17_6735053_6735295 | 0.10 |
TEKT1 |
tektin 1 |
94 |
0.96 |
| chr19_50935005_50935230 | 0.10 |
MYBPC2 |
myosin binding protein C, fast type |
1043 |
0.32 |
| chr8_71971726_71971877 | 0.10 |
RP11-326E22.1 |
|
95891 |
0.09 |
| chr16_20915372_20915539 | 0.10 |
LYRM1 |
LYR motif containing 1 |
3029 |
0.2 |
| chr3_184209968_184210530 | 0.10 |
EIF2B5-IT1 |
EIF2B5 intronic transcript 1 (non-protein coding) |
12652 |
0.18 |
| chr17_60896968_60897294 | 0.10 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
11426 |
0.25 |
| chr10_129718407_129718558 | 0.09 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
13066 |
0.2 |
| chr1_157013915_157014350 | 0.09 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
733 |
0.68 |
| chr7_138719583_138719734 | 0.09 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
1117 |
0.55 |
| chr9_100836589_100836740 | 0.09 |
NANS |
N-acetylneuraminic acid synthase |
2447 |
0.27 |
| chr2_150240204_150240355 | 0.09 |
LYPD6 |
LY6/PLAUR domain containing 6 |
16064 |
0.29 |
| chr3_150088154_150088521 | 0.09 |
TSC22D2 |
TSC22 domain family, member 2 |
37785 |
0.2 |
| chr15_38547811_38548278 | 0.09 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
2662 |
0.42 |
| chr15_63575130_63575281 | 0.09 |
APH1B |
APH1B gamma secretase subunit |
5388 |
0.27 |
| chr7_27187795_27188438 | 0.09 |
HOXA6 |
homeobox A6 |
723 |
0.31 |
| chr16_50721200_50721351 | 0.08 |
SNX20 |
sorting nexin 20 |
6011 |
0.13 |
| chr10_101743145_101743349 | 0.08 |
DNMBP |
dynamin binding protein |
26429 |
0.16 |
| chr2_178140372_178140777 | 0.08 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
10715 |
0.11 |
| chr4_182673763_182673929 | 0.08 |
ENSG00000251742 |
. |
82415 |
0.11 |
| chr8_42063953_42065062 | 0.08 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr5_74634021_74634424 | 0.08 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
264 |
0.89 |
| chr19_19496596_19496747 | 0.08 |
GATAD2A |
GATA zinc finger domain containing 2A |
29 |
0.97 |
| chr10_29010987_29011704 | 0.08 |
ENSG00000201001 |
. |
38387 |
0.14 |
| chr12_120672889_120673226 | 0.08 |
PXN |
paxillin |
8410 |
0.12 |
| chr2_203243816_203243967 | 0.08 |
BMPR2 |
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
1734 |
0.33 |
| chr6_44026023_44026284 | 0.08 |
RP5-1120P11.1 |
|
16236 |
0.16 |
| chr7_27191813_27192140 | 0.08 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
2305 |
0.1 |
| chr11_58730865_58731226 | 0.08 |
RP11-142C4.6 |
|
15713 |
0.15 |
| chr16_377314_377692 | 0.07 |
AXIN1 |
axin 1 |
24946 |
0.09 |
| chr9_135957875_135958026 | 0.07 |
CELP |
carboxyl ester lipase pseudogene |
24 |
0.96 |
| chr12_27939182_27939333 | 0.07 |
RP11-860B13.1 |
|
5257 |
0.13 |
| chr10_105436749_105436937 | 0.07 |
SH3PXD2A |
SH3 and PX domains 2A |
1079 |
0.5 |
| chr11_57121969_57122157 | 0.07 |
ENSG00000266018 |
. |
5133 |
0.1 |
| chr2_169963812_169963963 | 0.07 |
AC007556.3 |
|
6634 |
0.24 |
| chr6_52172375_52172526 | 0.07 |
MCM3 |
minichromosome maintenance complex component 3 |
22815 |
0.19 |
| chr12_75723842_75724113 | 0.07 |
CAPS2 |
calcyphosine 2 |
141 |
0.96 |
| chr5_39424296_39424549 | 0.07 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
548 |
0.85 |
| chr9_19999361_19999512 | 0.07 |
ENSG00000266224 |
. |
169685 |
0.03 |
| chr6_14743251_14743453 | 0.07 |
ENSG00000206960 |
. |
96586 |
0.09 |
| chr17_81021338_81021489 | 0.07 |
B3GNTL1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
11727 |
0.21 |
| chr5_57751053_57751204 | 0.07 |
PLK2 |
polo-like kinase 2 |
4959 |
0.26 |
| chr9_139513973_139514190 | 0.07 |
ENSG00000252440 |
. |
17114 |
0.1 |
| chr19_54600109_54600278 | 0.07 |
OSCAR |
osteoclast associated, immunoglobulin-like receptor |
3890 |
0.09 |
| chr2_233703105_233703293 | 0.06 |
GIGYF2 |
GRB10 interacting GYF protein 2 |
4471 |
0.17 |
| chr1_24529962_24530113 | 0.06 |
IFNLR1 |
interferon, lambda receptor 1 |
15588 |
0.16 |
| chr15_26273691_26273918 | 0.06 |
ENSG00000212604 |
. |
19583 |
0.26 |
| chr11_69213982_69214133 | 0.06 |
MYEOV |
myeloma overexpressed |
152432 |
0.04 |
| chr3_120067269_120067913 | 0.06 |
LRRC58 |
leucine rich repeat containing 58 |
595 |
0.53 |
| chr9_135937365_135937516 | 0.06 |
CEL |
carboxyl ester lipase |
75 |
0.96 |
| chr6_12291567_12292004 | 0.06 |
EDN1 |
endothelin 1 |
1189 |
0.64 |
| chr4_185270855_185271006 | 0.06 |
IRF2 |
interferon regulatory factor 2 |
68814 |
0.09 |
| chr16_2203805_2204060 | 0.06 |
RAB26 |
RAB26, member RAS oncogene family |
889 |
0.23 |
| chr4_139939633_139940037 | 0.06 |
ENSG00000264953 |
. |
1102 |
0.42 |
| chr11_34393219_34393432 | 0.06 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
13770 |
0.25 |
| chr20_37076011_37076413 | 0.06 |
ENSG00000174365 |
. |
515 |
0.58 |
| chr1_206831480_206831631 | 0.06 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
22381 |
0.12 |
| chr6_44279109_44279260 | 0.06 |
AARS2 |
alanyl-tRNA synthetase 2, mitochondrial |
1879 |
0.23 |
| chr10_104543017_104543364 | 0.06 |
WBP1L |
WW domain binding protein 1-like |
7184 |
0.15 |
| chr4_139328520_139328671 | 0.06 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
165092 |
0.04 |
| chr1_167408487_167409140 | 0.06 |
RP11-104L21.2 |
|
19085 |
0.19 |
| chr9_90115975_90116126 | 0.06 |
DAPK1 |
death-associated protein kinase 1 |
2130 |
0.42 |
| chr7_132299406_132299928 | 0.06 |
PLXNA4 |
plexin A4 |
33780 |
0.18 |
| chr7_92466339_92466990 | 0.06 |
CDK6 |
cyclin-dependent kinase 6 |
756 |
0.48 |
| chr17_1480739_1481169 | 0.05 |
PITPNA |
phosphatidylinositol transfer protein, alpha |
14844 |
0.11 |
| chr9_75566785_75567046 | 0.05 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
1056 |
0.7 |
| chr9_136602785_136602936 | 0.05 |
SARDH |
sarcosine dehydrogenase |
617 |
0.78 |
| chr2_143829603_143829754 | 0.05 |
ARHGAP15 |
Rho GTPase activating protein 15 |
19253 |
0.25 |
| chr12_125400700_125400851 | 0.05 |
ENSG00000265345 |
. |
682 |
0.48 |
| chr7_41735666_41735856 | 0.05 |
INHBA-AS1 |
INHBA antisense RNA 1 |
2215 |
0.31 |
| chr4_160150065_160150216 | 0.05 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
37893 |
0.17 |
| chr9_93577500_93577651 | 0.05 |
SYK |
spleen tyrosine kinase |
12195 |
0.32 |
| chr2_175198249_175198424 | 0.05 |
SP9 |
Sp9 transcription factor |
1338 |
0.39 |
| chr20_47897294_47897664 | 0.05 |
ENSG00000212304 |
. |
259 |
0.71 |
| chr14_69074893_69075044 | 0.05 |
CTD-2325P2.4 |
|
20194 |
0.24 |
| chr6_16683747_16684279 | 0.05 |
RP1-151F17.1 |
|
77356 |
0.11 |
| chr19_51013968_51014417 | 0.05 |
JOSD2 |
Josephin domain containing 2 |
153 |
0.91 |
| chr7_80929142_80929293 | 0.05 |
AC005008.2 |
Uncharacterized protein |
112111 |
0.07 |
| chr1_150480831_150481087 | 0.05 |
ECM1 |
extracellular matrix protein 1 |
376 |
0.77 |
| chr4_88939906_88940142 | 0.05 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
11204 |
0.19 |
| chr15_99092145_99092331 | 0.05 |
FAM169B |
family with sequence similarity 169, member B |
34627 |
0.21 |
| chr4_106631816_106631986 | 0.05 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
66 |
0.97 |
| chr19_15753132_15753283 | 0.05 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
1088 |
0.5 |
| chr1_181002765_181003143 | 0.05 |
MR1 |
major histocompatibility complex, class I-related |
113 |
0.96 |
| chr16_67564911_67565176 | 0.05 |
FAM65A |
family with sequence similarity 65, member A |
408 |
0.7 |
| chrX_115085542_115085858 | 0.05 |
ENSG00000266828 |
. |
23400 |
0.24 |
| chr6_138876582_138876733 | 0.05 |
NHSL1 |
NHS-like 1 |
9809 |
0.27 |
| chr18_55438742_55438893 | 0.05 |
ENSG00000202159 |
. |
16191 |
0.19 |
| chr11_86511248_86511444 | 0.05 |
PRSS23 |
protease, serine, 23 |
64 |
0.99 |
| chr1_47077616_47077767 | 0.05 |
MOB3C |
MOB kinase activator 3C |
3114 |
0.19 |
| chr6_114175848_114176208 | 0.05 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
2513 |
0.28 |
| chr3_53195990_53196532 | 0.05 |
PRKCD |
protein kinase C, delta |
724 |
0.65 |
| chr3_130568062_130568213 | 0.05 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
1302 |
0.48 |
| chr11_58349436_58349587 | 0.05 |
ZFP91 |
ZFP91 zinc finger protein |
2927 |
0.2 |
| chr20_11899797_11899948 | 0.05 |
BTBD3 |
BTB (POZ) domain containing 3 |
373 |
0.63 |
| chr10_129052216_129052367 | 0.05 |
FAM196A |
family with sequence similarity 196, member A |
57869 |
0.16 |
| chr20_3730561_3730712 | 0.04 |
C20orf27 |
chromosome 20 open reading frame 27 |
8689 |
0.12 |
| chr18_10131788_10132277 | 0.04 |
ENSG00000263630 |
. |
126838 |
0.05 |
| chr17_2293949_2294100 | 0.04 |
RP1-59D14.1 |
|
5883 |
0.11 |
| chr10_127754442_127754593 | 0.04 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
70531 |
0.1 |
| chr17_28246226_28246497 | 0.04 |
EFCAB5 |
EF-hand calcium binding domain 5 |
9857 |
0.15 |
| chr20_48989399_48989550 | 0.04 |
ENSG00000244376 |
. |
56546 |
0.13 |
| chr17_53829012_53829414 | 0.04 |
PCTP |
phosphatidylcholine transfer protein |
452 |
0.87 |
| chr9_97632930_97633081 | 0.04 |
RP11-49O14.2 |
|
29706 |
0.15 |
| chr11_121460609_121460760 | 0.04 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
444 |
0.91 |
| chr4_185235485_185235636 | 0.04 |
ENSG00000244512 |
. |
35471 |
0.16 |
| chr20_17660855_17661298 | 0.04 |
RRBP1 |
ribosome binding protein 1 |
1629 |
0.4 |
| chr7_106358902_106359239 | 0.04 |
CTB-111H14.1 |
|
13825 |
0.24 |
| chr6_88973705_88973856 | 0.04 |
CNR1 |
cannabinoid receptor 1 (brain) |
97702 |
0.09 |
| chr14_89493302_89493776 | 0.04 |
FOXN3 |
forkhead box N3 |
153549 |
0.04 |
| chr22_46443288_46443533 | 0.04 |
RP6-109B7.5 |
|
5563 |
0.11 |
| chr8_146053525_146053811 | 0.04 |
ZNF7 |
zinc finger protein 7 |
665 |
0.57 |
| chr16_10713609_10713760 | 0.04 |
EMP2 |
epithelial membrane protein 2 |
39129 |
0.13 |
| chr15_86076847_86076998 | 0.04 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
10349 |
0.21 |
| chr17_60501297_60502176 | 0.04 |
METTL2A |
methyltransferase like 2A |
508 |
0.79 |
| chr6_119069398_119069957 | 0.04 |
CEP85L |
centrosomal protein 85kDa-like |
38439 |
0.19 |
| chr9_85834043_85834730 | 0.04 |
RP11-439K3.1 |
|
140 |
0.96 |
| chr2_24232585_24233394 | 0.04 |
MFSD2B |
major facilitator superfamily domain containing 2B |
22 |
0.97 |
| chr5_10306978_10307734 | 0.04 |
CMBL |
carboxymethylenebutenolidase homolog (Pseudomonas) |
782 |
0.53 |
| chr1_68153534_68153824 | 0.04 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
2795 |
0.31 |
| chr18_70535912_70536084 | 0.04 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
814 |
0.7 |
| chr2_68954923_68955084 | 0.04 |
ARHGAP25 |
Rho GTPase activating protein 25 |
6910 |
0.27 |
| chr5_115297082_115297364 | 0.04 |
AQPEP |
Aminopeptidase Q |
928 |
0.59 |
| chr17_15732358_15732519 | 0.04 |
AC015922.5 |
|
245 |
0.93 |
| chr2_192711472_192711842 | 0.04 |
SDPR |
serum deprivation response |
324 |
0.55 |
| chr3_188389672_188389823 | 0.04 |
ENSG00000207651 |
. |
16822 |
0.22 |
| chr7_40591526_40591750 | 0.04 |
AC004988.1 |
|
5111 |
0.35 |
| chr22_31626244_31626468 | 0.04 |
ENSG00000202019 |
. |
198 |
0.9 |
| chr2_48625526_48625677 | 0.04 |
PPP1R21 |
protein phosphatase 1, regulatory subunit 21 |
42307 |
0.15 |
| chr15_62798995_62799233 | 0.04 |
TLN2 |
talin 2 |
54450 |
0.15 |
| chr13_92759091_92759242 | 0.04 |
ENSG00000252508 |
. |
47069 |
0.2 |
| chr6_7911720_7911871 | 0.04 |
TXNDC5 |
thioredoxin domain containing 5 (endoplasmic reticulum) |
748 |
0.79 |
| chr3_128152097_128152248 | 0.04 |
DNAJB8-AS1 |
DNAJB8 antisense RNA 1 |
30265 |
0.14 |
| chr17_26649609_26649760 | 0.04 |
TMEM97 |
transmembrane protein 97 |
3203 |
0.1 |
| chr7_85101266_85101421 | 0.04 |
SEMA3D |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
285172 |
0.01 |
| chr12_104613717_104613915 | 0.04 |
TXNRD1 |
thioredoxin reductase 1 |
4257 |
0.25 |
| chr8_133333355_133333734 | 0.04 |
KCNQ3 |
potassium voltage-gated channel, KQT-like subfamily, member 3 |
125965 |
0.06 |
| chr16_89003315_89003466 | 0.04 |
RP11-830F9.7 |
|
774 |
0.51 |
| chr22_30268427_30268578 | 0.04 |
MTMR3 |
myotubularin related protein 3 |
10642 |
0.16 |
| chr19_2079674_2079825 | 0.04 |
MOB3A |
MOB kinase activator 3A |
1031 |
0.38 |
| chr10_94448695_94449449 | 0.04 |
HHEX |
hematopoietically expressed homeobox |
1127 |
0.51 |
| chr17_18575650_18575928 | 0.04 |
RP11-815I9.4 |
|
3015 |
0.17 |
| chr12_94207424_94207575 | 0.04 |
ENSG00000264978 |
. |
17353 |
0.19 |
| chr1_79472541_79472704 | 0.04 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
219 |
0.97 |
| chr9_36194632_36194925 | 0.04 |
CLTA |
clathrin, light chain A |
3818 |
0.22 |
| chr11_94509014_94509165 | 0.04 |
AMOTL1 |
angiomotin like 1 |
7552 |
0.24 |
| chr3_152122945_152123096 | 0.04 |
MBNL1 |
muscleblind-like splicing regulator 1 |
9827 |
0.23 |
| chr9_92724401_92724683 | 0.04 |
ENSG00000263967 |
. |
61275 |
0.16 |
| chr5_173959397_173959548 | 0.04 |
MSX2 |
msh homeobox 2 |
192064 |
0.03 |
| chr5_100113349_100113842 | 0.04 |
ENSG00000221263 |
. |
38674 |
0.19 |
| chr7_22762779_22763152 | 0.04 |
IL6 |
interleukin 6 (interferon, beta 2) |
2538 |
0.27 |
| chr16_78344596_78344747 | 0.04 |
RP11-190D6.2 |
|
69556 |
0.13 |
| chr12_32687185_32687743 | 0.04 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
206 |
0.96 |
| chr7_129933276_129933885 | 0.04 |
CPA4 |
carboxypeptidase A4 |
461 |
0.76 |
| chr7_116167584_116167883 | 0.04 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
1386 |
0.37 |
| chr11_13238316_13238540 | 0.04 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
59771 |
0.15 |
| chr9_110495286_110496024 | 0.03 |
AL162389.1 |
Uncharacterized protein |
44764 |
0.15 |
| chr11_2847450_2848020 | 0.03 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
35063 |
0.12 |
| chr22_30600911_30601062 | 0.03 |
RP3-438O4.4 |
|
2112 |
0.23 |
| chr16_88923128_88923365 | 0.03 |
GALNS |
galactosamine (N-acetyl)-6-sulfate sulfatase |
39 |
0.75 |
| chr7_557703_557916 | 0.03 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
336 |
0.89 |
| chr6_18260761_18260912 | 0.03 |
DEK |
DEK oncogene |
3603 |
0.23 |
| chr6_119255807_119256176 | 0.03 |
RP11-351A11.1 |
|
41 |
0.71 |
| chr1_204983840_204983991 | 0.03 |
NFASC |
neurofascin |
17603 |
0.17 |
| chr8_145747471_145747843 | 0.03 |
LRRC14 |
leucine rich repeat containing 14 |
3528 |
0.08 |
| chr7_73624408_73624729 | 0.03 |
LAT2 |
linker for activation of T cells family, member 2 |
215 |
0.92 |
| chr12_109227226_109227475 | 0.03 |
ENSG00000207622 |
. |
3432 |
0.19 |
| chr10_121419548_121419699 | 0.03 |
BAG3 |
BCL2-associated athanogene 3 |
3501 |
0.28 |
| chr20_16031217_16031368 | 0.03 |
MACROD2 |
MACRO domain containing 2 |
64859 |
0.15 |
| chr3_44063258_44064101 | 0.03 |
ENSG00000252980 |
. |
48900 |
0.17 |
| chr12_31515445_31515828 | 0.03 |
ENSG00000207477 |
. |
3586 |
0.2 |
| chr5_135527961_135528159 | 0.03 |
TRPC7-AS1 |
TRPC7 antisense RNA 1 |
21676 |
0.18 |
| chr5_94957151_94957390 | 0.03 |
GPR150 |
G protein-coupled receptor 150 |
1488 |
0.38 |
| chr12_79848122_79848556 | 0.03 |
ENSG00000266390 |
. |
7991 |
0.2 |
| chr6_24666286_24667027 | 0.03 |
TDP2 |
tyrosyl-DNA phosphodiesterase 2 |
163 |
0.84 |
| chr14_89833069_89833220 | 0.03 |
RP11-356K23.2 |
|
11740 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |