Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
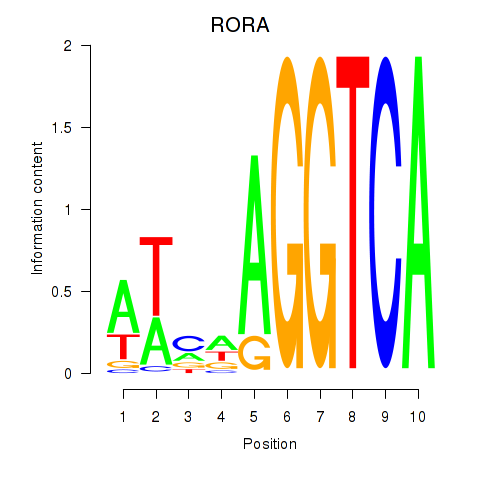
Results for RORA
Z-value: 0.71
Transcription factors associated with RORA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORA
|
ENSG00000069667.11 | RAR related orphan receptor A |
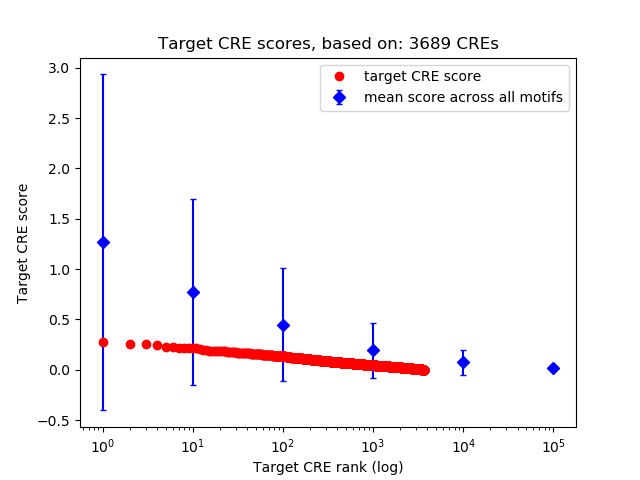
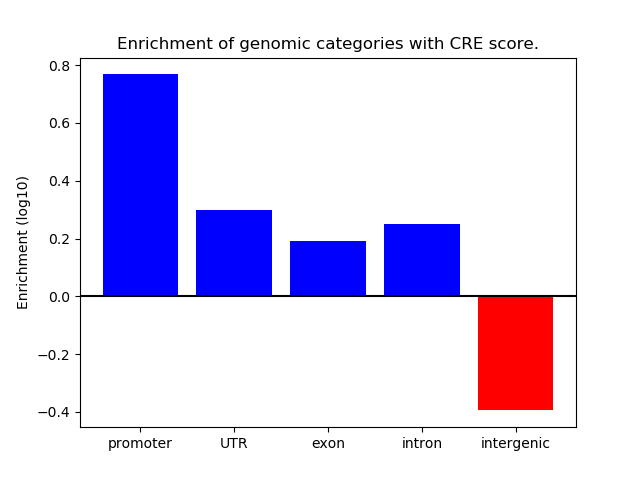
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_61611314_61611465 | RORA | 89871 | 0.090299 | 0.72 | 2.8e-02 | Click! |
| chr15_61521269_61521446 | RORA | 161 | 0.963755 | 0.52 | 1.5e-01 | Click! |
| chr15_60878348_60878499 | RORA | 6317 | 0.239541 | -0.48 | 1.9e-01 | Click! |
| chr15_60897779_60897930 | RORA | 12529 | 0.226573 | 0.48 | 1.9e-01 | Click! |
| chr15_60881361_60881613 | RORA | 3253 | 0.291829 | -0.47 | 2.1e-01 | Click! |
Activity of the RORA motif across conditions
Conditions sorted by the z-value of the RORA motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
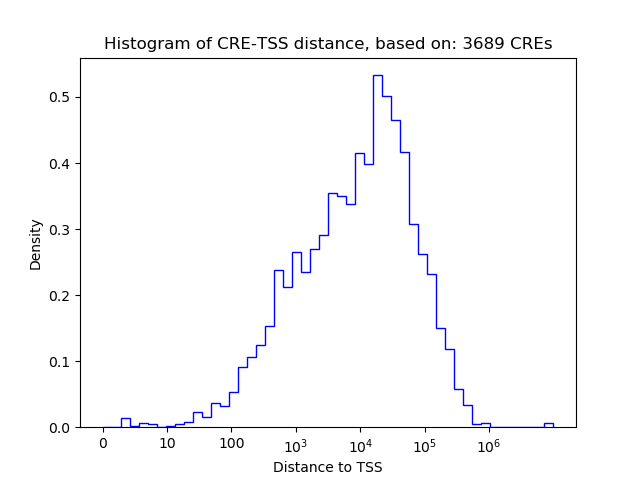
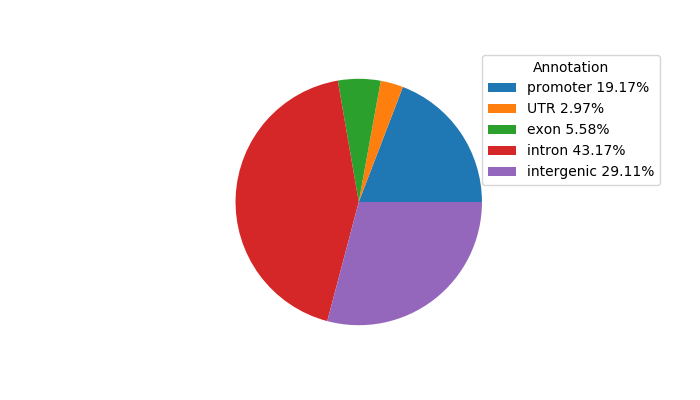
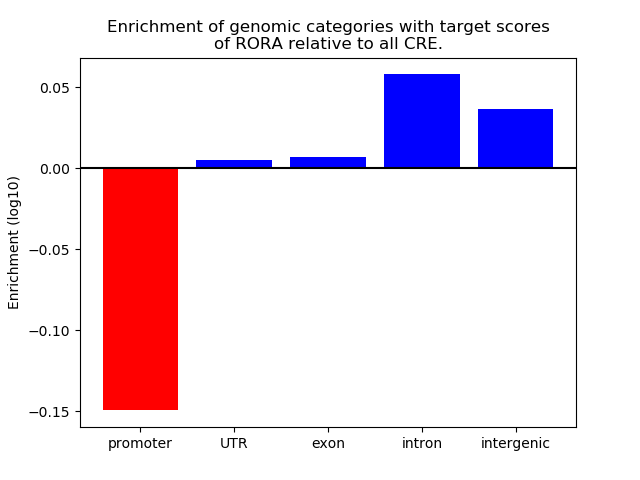
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_6567467_6567618 | 0.28 |
ENSG00000252110 |
. |
1117 |
0.53 |
| chr3_184037306_184037783 | 0.26 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
1 |
0.95 |
| chr11_132813237_132813595 | 0.25 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
150 |
0.98 |
| chr16_49677980_49678248 | 0.24 |
ZNF423 |
zinc finger protein 423 |
3351 |
0.32 |
| chrX_102940107_102940387 | 0.23 |
MORF4L2 |
mortality factor 4 like 2 |
902 |
0.43 |
| chr1_223302857_223303051 | 0.23 |
TLR5 |
toll-like receptor 5 |
5144 |
0.33 |
| chr5_39423074_39423254 | 0.22 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
1806 |
0.48 |
| chr8_50968595_50968898 | 0.21 |
SNTG1 |
syntrophin, gamma 1 |
144041 |
0.05 |
| chr4_89976890_89977204 | 0.21 |
FAM13A |
family with sequence similarity 13, member A |
1264 |
0.56 |
| chr12_50611414_50611565 | 0.21 |
RP3-405J10.4 |
|
1898 |
0.22 |
| chr7_101525780_101526170 | 0.21 |
CTA-339C12.1 |
|
57966 |
0.12 |
| chr2_243030793_243031816 | 0.20 |
AC093642.5 |
|
460 |
0.62 |
| chr4_176568169_176568320 | 0.20 |
GPM6A |
glycoprotein M6A |
140294 |
0.05 |
| chr4_183047320_183047598 | 0.20 |
AC108142.1 |
|
17470 |
0.17 |
| chr2_41837870_41838465 | 0.19 |
ENSG00000221372 |
. |
14641 |
0.29 |
| chr13_40976108_40976619 | 0.19 |
ENSG00000252812 |
. |
5214 |
0.32 |
| chr13_24006860_24007105 | 0.19 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
859 |
0.74 |
| chr17_3379575_3379800 | 0.19 |
ASPA |
aspartoacylase |
391 |
0.8 |
| chr7_92242274_92242425 | 0.19 |
FAM133B |
family with sequence similarity 133, member B |
22641 |
0.2 |
| chr17_65525390_65525554 | 0.19 |
CTD-2653B5.1 |
|
4875 |
0.23 |
| chr17_39265148_39265305 | 0.19 |
KRTAP4-9 |
keratin associated protein 4-9 |
3642 |
0.08 |
| chr3_20616495_20616647 | 0.19 |
ENSG00000206807 |
. |
67323 |
0.14 |
| chr6_27205685_27206711 | 0.18 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chr2_145218006_145218157 | 0.18 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
29944 |
0.22 |
| chr6_76147146_76147428 | 0.18 |
ENSG00000263533 |
. |
9164 |
0.18 |
| chr1_245316415_245317636 | 0.18 |
KIF26B |
kinesin family member 26B |
1262 |
0.47 |
| chr1_23610259_23610426 | 0.18 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
60413 |
0.09 |
| chr12_77748772_77748964 | 0.17 |
ENSG00000238769 |
. |
192063 |
0.03 |
| chr10_25007047_25007198 | 0.17 |
ARHGAP21 |
Rho GTPase activating protein 21 |
3673 |
0.34 |
| chr16_2517636_2518674 | 0.17 |
RP11-715J22.2 |
|
59 |
0.92 |
| chr3_183005602_183005777 | 0.17 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
22557 |
0.18 |
| chrX_100334578_100334939 | 0.17 |
TMEM35 |
transmembrane protein 35 |
1049 |
0.47 |
| chr6_86113632_86113933 | 0.17 |
NT5E |
5'-nucleotidase, ecto (CD73) |
46027 |
0.19 |
| chr4_77483255_77483500 | 0.17 |
ENSG00000263445 |
. |
11344 |
0.16 |
| chr7_101579659_101580126 | 0.17 |
CTB-181H17.1 |
|
23504 |
0.22 |
| chr8_27492286_27492503 | 0.17 |
SCARA3 |
scavenger receptor class A, member 3 |
696 |
0.66 |
| chr16_86700893_86701044 | 0.17 |
FOXL1 |
forkhead box L1 |
88853 |
0.08 |
| chr1_101359369_101359680 | 0.17 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
696 |
0.48 |
| chr1_96354702_96354941 | 0.16 |
ENSG00000221798 |
. |
2725 |
0.41 |
| chr7_4722786_4723588 | 0.16 |
FOXK1 |
forkhead box K1 |
1247 |
0.49 |
| chr14_93701571_93701751 | 0.16 |
UBR7 |
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
16634 |
0.14 |
| chrX_11456012_11456286 | 0.16 |
ARHGAP6 |
Rho GTPase activating protein 6 |
10256 |
0.3 |
| chr3_114161410_114161561 | 0.16 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
12045 |
0.25 |
| chr8_71580499_71580692 | 0.16 |
LACTB2 |
lactamase, beta 2 |
797 |
0.44 |
| chr2_190477763_190477914 | 0.16 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
29354 |
0.16 |
| chr16_65151579_65151789 | 0.16 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
4149 |
0.39 |
| chr8_122651204_122651355 | 0.16 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
254 |
0.93 |
| chr17_45302890_45303160 | 0.16 |
ENSG00000252088 |
. |
5017 |
0.14 |
| chr12_83211442_83211593 | 0.16 |
TMTC2 |
transmembrane and tetratricopeptide repeat containing 2 |
59183 |
0.17 |
| chr7_115116914_115117103 | 0.16 |
ENSG00000202377 |
. |
104488 |
0.08 |
| chr11_62162542_62162710 | 0.16 |
CTD-2531D15.5 |
|
1782 |
0.23 |
| chr6_140029993_140030144 | 0.15 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
334311 |
0.01 |
| chr1_90464057_90464208 | 0.15 |
ZNF326 |
zinc finger protein 326 |
3405 |
0.29 |
| chr6_116571521_116571672 | 0.15 |
TSPYL4 |
TSPY-like 4 |
3665 |
0.18 |
| chr22_46423027_46423794 | 0.15 |
RP6-109B7.5 |
|
25563 |
0.09 |
| chr3_143363258_143363409 | 0.15 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
203967 |
0.03 |
| chr3_189306684_189306989 | 0.15 |
TP63 |
tumor protein p63 |
42380 |
0.21 |
| chrX_51636554_51637686 | 0.15 |
MAGED1 |
melanoma antigen family D, 1 |
385 |
0.89 |
| chr1_78806002_78806206 | 0.15 |
ENSG00000212308 |
. |
34849 |
0.18 |
| chr2_46173551_46173702 | 0.15 |
PRKCE |
protein kinase C, epsilon |
54415 |
0.16 |
| chr1_244836322_244836473 | 0.15 |
DESI2 |
desumoylating isopeptidase 2 |
19344 |
0.23 |
| chr6_84139999_84140706 | 0.15 |
ME1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
412 |
0.89 |
| chr14_104023687_104023838 | 0.15 |
RP11-894P9.2 |
|
4004 |
0.09 |
| chr5_36302348_36302599 | 0.15 |
RANBP3L |
RAN binding protein 3-like |
257 |
0.94 |
| chrX_3263158_3263429 | 0.15 |
MXRA5 |
matrix-remodelling associated 5 |
1389 |
0.55 |
| chr2_197957408_197957559 | 0.15 |
ANKRD44 |
ankyrin repeat domain 44 |
18085 |
0.25 |
| chr2_237444042_237444193 | 0.15 |
IQCA1 |
IQ motif containing with AAA domain 1 |
27932 |
0.2 |
| chr6_143508899_143509050 | 0.14 |
AIG1 |
androgen-induced 1 |
61587 |
0.13 |
| chr7_95545859_95546042 | 0.14 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
112354 |
0.07 |
| chr2_66672542_66672972 | 0.14 |
MEIS1 |
Meis homeobox 1 |
2660 |
0.22 |
| chr21_33516188_33516339 | 0.14 |
ENSG00000264055 |
. |
7954 |
0.26 |
| chr17_4981225_4982338 | 0.14 |
ZFP3 |
ZFP3 zinc finger protein |
238 |
0.6 |
| chr6_80520238_80520389 | 0.14 |
ENSG00000221470 |
. |
16289 |
0.28 |
| chr2_190272487_190272871 | 0.14 |
WDR75 |
WD repeat domain 75 |
33480 |
0.21 |
| chr19_57067071_57067222 | 0.14 |
AC007228.11 |
|
11634 |
0.12 |
| chr8_57594130_57594603 | 0.14 |
RP11-17A4.2 |
|
192709 |
0.03 |
| chr17_29543083_29543234 | 0.14 |
NF1 |
neurofibromin 1 |
15604 |
0.18 |
| chr10_54074428_54074776 | 0.14 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
546 |
0.57 |
| chr17_67651034_67651185 | 0.14 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
152539 |
0.04 |
| chr19_34359259_34359976 | 0.14 |
ENSG00000240626 |
. |
58913 |
0.13 |
| chr3_149271657_149271885 | 0.14 |
WWTR1 |
WW domain containing transcription regulator 1 |
22268 |
0.17 |
| chr20_53092813_53093232 | 0.14 |
DOK5 |
docking protein 5 |
765 |
0.8 |
| chr13_20175736_20176974 | 0.14 |
MPHOSPH8 |
M-phase phosphoprotein 8 |
31433 |
0.18 |
| chr16_25022284_25022579 | 0.14 |
ARHGAP17 |
Rho GTPase activating protein 17 |
4221 |
0.31 |
| chr5_87436294_87437103 | 0.14 |
TMEM161B |
transmembrane protein 161B |
79750 |
0.11 |
| chr2_101435204_101435829 | 0.14 |
NPAS2 |
neuronal PAS domain protein 2 |
1098 |
0.54 |
| chr5_112417183_112417334 | 0.14 |
DCP2 |
decapping mRNA 2 |
104779 |
0.06 |
| chr6_148685677_148685874 | 0.14 |
SASH1 |
SAM and SH3 domain containing 1 |
22046 |
0.24 |
| chr10_29935760_29936003 | 0.14 |
SVIL |
supervillin |
11980 |
0.21 |
| chr4_81192880_81193031 | 0.14 |
FGF5 |
fibroblast growth factor 5 |
5162 |
0.27 |
| chr10_62588054_62588205 | 0.14 |
CDK1 |
cyclin-dependent kinase 1 |
48218 |
0.16 |
| chr11_65199714_65199921 | 0.14 |
ENSG00000245532 |
. |
12112 |
0.11 |
| chr15_74232964_74233156 | 0.14 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
12471 |
0.14 |
| chr9_135895662_135895884 | 0.14 |
GTF3C5 |
general transcription factor IIIC, polypeptide 5, 63kDa |
10303 |
0.13 |
| chr5_142005239_142005585 | 0.14 |
FGF1 |
fibroblast growth factor 1 (acidic) |
4505 |
0.24 |
| chr6_56239918_56240091 | 0.14 |
COL21A1 |
collagen, type XXI, alpha 1 |
18888 |
0.21 |
| chr11_19736431_19736825 | 0.14 |
NAV2 |
neuron navigator 2 |
1485 |
0.49 |
| chr17_2394357_2394508 | 0.13 |
METTL16 |
methyltransferase like 16 |
20748 |
0.15 |
| chr18_72843553_72843775 | 0.13 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
73804 |
0.12 |
| chr8_136478593_136478847 | 0.13 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
7770 |
0.25 |
| chr4_37668617_37668785 | 0.13 |
RELL1 |
RELT-like 1 |
1698 |
0.42 |
| chr9_118378371_118378558 | 0.13 |
DEC1 |
deleted in esophageal cancer 1 |
474367 |
0.01 |
| chr11_100800789_100801024 | 0.13 |
ENSG00000238388 |
. |
4099 |
0.25 |
| chr9_678099_678250 | 0.13 |
RP11-130C19.3 |
|
546 |
0.81 |
| chr6_24743331_24743534 | 0.13 |
C6orf62 |
chromosome 6 open reading frame 62 |
22368 |
0.12 |
| chr3_72445670_72445821 | 0.13 |
RYBP |
RING1 and YY1 binding protein |
50324 |
0.17 |
| chr3_99591567_99591782 | 0.13 |
FILIP1L |
filamin A interacting protein 1-like |
3274 |
0.32 |
| chr18_479598_479768 | 0.13 |
COLEC12 |
collectin sub-family member 12 |
21039 |
0.18 |
| chr10_17275327_17275794 | 0.13 |
RP11-124N14.3 |
|
1272 |
0.37 |
| chr7_98248959_98249425 | 0.13 |
NPTX2 |
neuronal pentraxin II |
2583 |
0.39 |
| chr8_29133372_29133712 | 0.13 |
ENSG00000212034 |
. |
5964 |
0.17 |
| chr1_203503529_203503721 | 0.13 |
OPTC |
opticin |
38527 |
0.16 |
| chr11_132831774_132832000 | 0.13 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
18224 |
0.28 |
| chr1_161992740_161992891 | 0.13 |
OLFML2B |
olfactomedin-like 2B |
615 |
0.79 |
| chr3_187989024_187989175 | 0.12 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
31447 |
0.22 |
| chr5_88178054_88178448 | 0.12 |
MEF2C |
myocyte enhancer factor 2C |
713 |
0.52 |
| chr1_219116097_219116279 | 0.12 |
LYPLAL1 |
lysophospholipase-like 1 |
230998 |
0.02 |
| chr18_7568558_7569022 | 0.12 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
973 |
0.7 |
| chr8_8153665_8153908 | 0.12 |
ALG1L13P |
asparagine-linked glycosylation 1-like 13, pseudogene |
54853 |
0.11 |
| chr10_4283577_4283877 | 0.12 |
ENSG00000207124 |
. |
273417 |
0.02 |
| chr9_693371_693572 | 0.12 |
RP11-130C19.3 |
|
7916 |
0.22 |
| chr16_70749819_70749970 | 0.12 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
15746 |
0.13 |
| chr3_157078098_157078249 | 0.12 |
ENSG00000243176 |
. |
74224 |
0.1 |
| chr5_67825283_67825434 | 0.12 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
236962 |
0.02 |
| chr1_41176173_41176466 | 0.12 |
NFYC |
nuclear transcription factor Y, gamma |
1104 |
0.44 |
| chr6_89692667_89692818 | 0.12 |
AL079342.1 |
Uncharacterized protein; cDNA FLJ27030 fis, clone SLV07741 |
18496 |
0.18 |
| chr7_27217405_27217623 | 0.12 |
RP1-170O19.20 |
Uncharacterized protein |
2118 |
0.1 |
| chr13_113841496_113841673 | 0.12 |
PCID2 |
PCI domain containing 2 |
20836 |
0.11 |
| chr18_77746974_77747125 | 0.12 |
TXNL4A |
thioredoxin-like 4A |
1488 |
0.39 |
| chr2_236401822_236402641 | 0.12 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
502 |
0.83 |
| chr4_147568677_147568828 | 0.12 |
ENSG00000264323 |
. |
8339 |
0.22 |
| chr15_99440572_99440803 | 0.12 |
RP11-654A16.1 |
|
3923 |
0.25 |
| chr12_56241387_56241673 | 0.12 |
MMP19 |
matrix metallopeptidase 19 |
4796 |
0.11 |
| chr10_116527243_116527917 | 0.12 |
FAM160B1 |
family with sequence similarity 160, member B1 |
53923 |
0.15 |
| chr4_41215270_41215769 | 0.12 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
956 |
0.59 |
| chr3_89153572_89153739 | 0.12 |
EPHA3 |
EPH receptor A3 |
3019 |
0.42 |
| chr1_205326238_205326434 | 0.12 |
KLHDC8A |
kelch domain containing 8A |
118 |
0.96 |
| chr6_167764253_167765165 | 0.12 |
TTLL2 |
tubulin tyrosine ligase-like family, member 2 |
26135 |
0.18 |
| chr13_76026824_76027007 | 0.12 |
TBC1D4 |
TBC1 domain family, member 4 |
29335 |
0.19 |
| chr11_7715663_7715870 | 0.12 |
RP11-35J10.4 |
|
11069 |
0.15 |
| chr14_55119300_55119828 | 0.12 |
SAMD4A |
sterile alpha motif domain containing 4A |
84927 |
0.09 |
| chr14_42072367_42072518 | 0.12 |
LRFN5 |
leucine rich repeat and fibronectin type III domain containing 5 |
4331 |
0.28 |
| chr8_123867992_123868238 | 0.12 |
ZHX2 |
zinc fingers and homeoboxes 2 |
7509 |
0.2 |
| chr12_10868992_10869331 | 0.12 |
YBX3 |
Y box binding protein 3 |
6745 |
0.18 |
| chr6_53928343_53928541 | 0.12 |
MLIP-AS1 |
MLIP antisense RNA 1 |
16050 |
0.2 |
| chr6_116726836_116726987 | 0.12 |
DSE |
dermatan sulfate epimerase |
34801 |
0.12 |
| chr8_19148668_19149118 | 0.12 |
SH2D4A |
SH2 domain containing 4A |
22235 |
0.28 |
| chr3_73505367_73505518 | 0.12 |
PDZRN3 |
PDZ domain containing ring finger 3 |
18341 |
0.27 |
| chr1_234630809_234631231 | 0.11 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
16171 |
0.21 |
| chr1_31883502_31883784 | 0.11 |
SERINC2 |
serine incorporator 2 |
595 |
0.71 |
| chr2_45004607_45004758 | 0.11 |
ENSG00000252896 |
. |
4587 |
0.34 |
| chr15_100583304_100583570 | 0.11 |
ENSG00000252957 |
. |
30958 |
0.16 |
| chr7_130595711_130595921 | 0.11 |
ENSG00000226380 |
. |
33518 |
0.2 |
| chrX_135267737_135267902 | 0.11 |
FHL1 |
four and a half LIM domains 1 |
11094 |
0.22 |
| chr3_66101350_66101501 | 0.11 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
17860 |
0.25 |
| chr3_37352486_37352637 | 0.11 |
GOLGA4 |
golgin A4 |
28887 |
0.15 |
| chr2_143919802_143919953 | 0.11 |
RP11-190J23.1 |
|
9864 |
0.26 |
| chr3_21686525_21686676 | 0.11 |
ZNF385D-AS1 |
ZNF385D antisense RNA 1 |
102292 |
0.08 |
| chr2_216298275_216298426 | 0.11 |
FN1 |
fibronectin 1 |
2440 |
0.3 |
| chr1_150480597_150480804 | 0.11 |
ECM1 |
extracellular matrix protein 1 |
117 |
0.94 |
| chr19_55996251_55997143 | 0.11 |
NAT14 |
N-acetyltransferase 14 (GCN5-related, putative) |
96 |
0.92 |
| chr9_21558481_21558945 | 0.11 |
MIR31HG |
MIR31 host gene (non-protein coding) |
955 |
0.56 |
| chr18_40710046_40710304 | 0.11 |
RIT2 |
Ras-like without CAAX 2 |
14518 |
0.31 |
| chr2_227658957_227659437 | 0.11 |
IRS1 |
insulin receptor substrate 1 |
5278 |
0.23 |
| chr11_102340087_102340238 | 0.11 |
RP11-315O6.2 |
HCG1815860; Uncharacterized protein |
2176 |
0.27 |
| chr4_169615079_169615336 | 0.11 |
ENSG00000206613 |
. |
7340 |
0.22 |
| chr6_170053828_170054109 | 0.11 |
WDR27 |
WD repeat domain 27 |
6820 |
0.18 |
| chr10_15476627_15476827 | 0.11 |
FAM171A1 |
family with sequence similarity 171, member A1 |
63666 |
0.14 |
| chr3_124420440_124420591 | 0.11 |
ENSG00000263775 |
. |
4632 |
0.17 |
| chr2_20795534_20795685 | 0.11 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
3301 |
0.28 |
| chr11_26353711_26353957 | 0.11 |
ANO3 |
anoctamin 3 |
166 |
0.98 |
| chr8_20186622_20186773 | 0.11 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
25223 |
0.2 |
| chr5_102862275_102862460 | 0.11 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
36123 |
0.24 |
| chr15_83501556_83501772 | 0.11 |
HOMER2 |
homer homolog 2 (Drosophila) |
16815 |
0.13 |
| chr3_156409051_156409287 | 0.11 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
10214 |
0.23 |
| chr11_121835249_121835400 | 0.11 |
ENSG00000252556 |
. |
39739 |
0.19 |
| chrX_120775735_120775886 | 0.11 |
ENSG00000221217 |
. |
226910 |
0.02 |
| chr1_160311738_160311889 | 0.11 |
COPA |
coatomer protein complex, subunit alpha |
1225 |
0.26 |
| chr3_54462200_54462389 | 0.11 |
ESRG |
embryonic stem cell related (non-protein coding) |
211590 |
0.02 |
| chr7_94029074_94029225 | 0.11 |
COL1A2 |
collagen, type I, alpha 2 |
5276 |
0.3 |
| chr21_16663113_16663407 | 0.11 |
NRIP1 |
nuclear receptor interacting protein 1 |
225939 |
0.02 |
| chr6_116382044_116382245 | 0.11 |
FRK |
fyn-related kinase |
223 |
0.94 |
| chr2_7663684_7663835 | 0.11 |
ENSG00000221255 |
. |
53213 |
0.19 |
| chr2_190017431_190017582 | 0.11 |
ENSG00000264725 |
. |
19669 |
0.19 |
| chr6_24901738_24901889 | 0.11 |
FAM65B |
family with sequence similarity 65, member B |
9382 |
0.22 |
| chrX_99899594_99899808 | 0.11 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
486 |
0.79 |
| chr4_145568031_145568374 | 0.11 |
HHIP |
hedgehog interacting protein |
878 |
0.49 |
| chr17_13265618_13265967 | 0.11 |
ENSG00000266115 |
. |
137853 |
0.05 |
| chr6_27462859_27463076 | 0.11 |
ZNF184 |
zinc finger protein 184 |
22070 |
0.21 |
| chr4_157872609_157873145 | 0.11 |
PDGFC |
platelet derived growth factor C |
19178 |
0.21 |
| chr10_14920855_14921866 | 0.11 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
439 |
0.82 |
| chr13_60709592_60709946 | 0.11 |
DIAPH3-AS2 |
DIAPH3 antisense RNA 2 |
9063 |
0.22 |
| chr6_139691315_139691533 | 0.11 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
3926 |
0.31 |
| chr5_103849207_103849358 | 0.11 |
ENSG00000239808 |
. |
252755 |
0.02 |
| chrX_100545955_100546357 | 0.10 |
TAF7L |
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
168 |
0.93 |
| chr9_128000677_128001068 | 0.10 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
2737 |
0.2 |
| chr7_32627060_32627248 | 0.10 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
44287 |
0.18 |
| chr8_106487505_106487656 | 0.10 |
ZFPM2 |
zinc finger protein, FOG family member 2 |
53532 |
0.18 |
| chr4_88786510_88786661 | 0.10 |
MEPE |
matrix extracellular phosphoglycoprotein |
32446 |
0.16 |
| chr6_27516181_27516437 | 0.10 |
ENSG00000206671 |
. |
47882 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0052308 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0045652 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |