Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
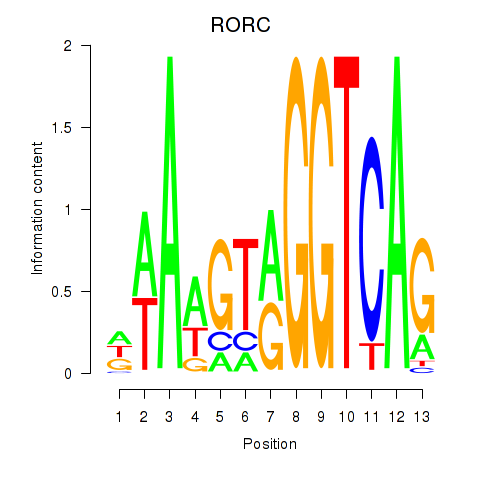
Results for RORC
Z-value: 0.82
Transcription factors associated with RORC
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RORC
|
ENSG00000143365.12 | RAR related orphan receptor C |
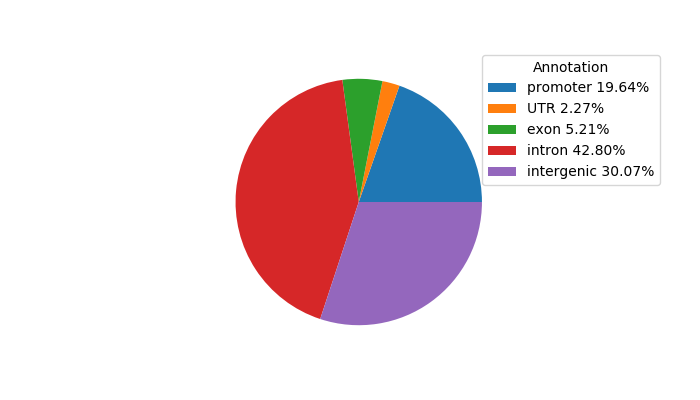
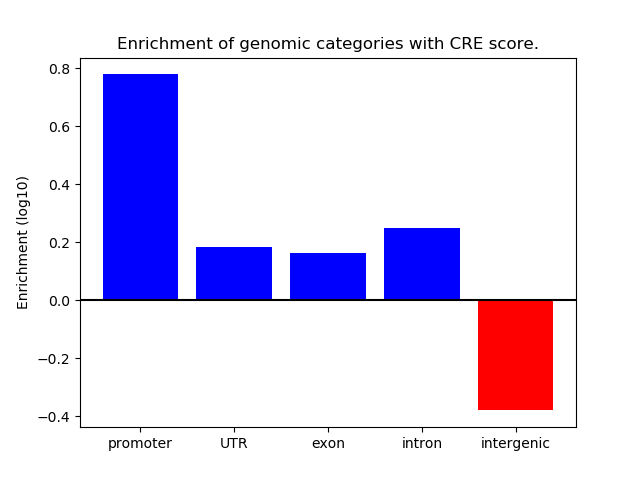
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_151796458_151796609 | RORC | 2034 | 0.141495 | -0.84 | 5.0e-03 | Click! |
| chr1_151797825_151797976 | RORC | 667 | 0.457926 | -0.66 | 5.3e-02 | Click! |
| chr1_151798177_151798381 | RORC | 288 | 0.776551 | -0.63 | 6.7e-02 | Click! |
| chr1_151795357_151795508 | RORC | 3135 | 0.102930 | -0.63 | 7.1e-02 | Click! |
| chr1_151796793_151796944 | RORC | 1699 | 0.167965 | -0.55 | 1.2e-01 | Click! |
Activity of the RORC motif across conditions
Conditions sorted by the z-value of the RORC motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
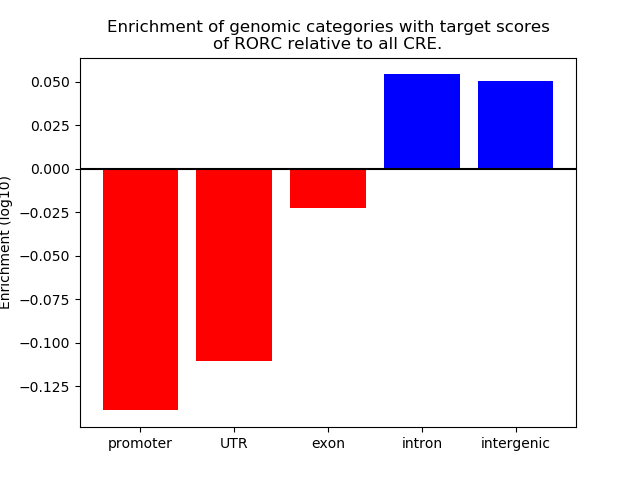
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_55996251_55997143 | 0.54 |
NAT14 |
N-acetyltransferase 14 (GCN5-related, putative) |
96 |
0.92 |
| chr4_71570546_71572213 | 0.45 |
RUFY3 |
RUN and FYVE domain containing 3 |
861 |
0.36 |
| chr15_72529055_72529849 | 0.39 |
PKM |
pyruvate kinase, muscle |
5288 |
0.17 |
| chr15_31507532_31508310 | 0.34 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
54445 |
0.15 |
| chr12_81258392_81258564 | 0.33 |
LIN7A |
lin-7 homolog A (C. elegans) |
24649 |
0.18 |
| chr10_30092462_30092778 | 0.32 |
SVIL |
supervillin |
67887 |
0.12 |
| chr8_128957071_128957260 | 0.30 |
TMEM75 |
transmembrane protein 75 |
3426 |
0.25 |
| chr9_94180104_94180255 | 0.30 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
5965 |
0.3 |
| chr10_3929254_3929732 | 0.29 |
KLF6 |
Kruppel-like factor 6 |
102020 |
0.08 |
| chr7_29603979_29604650 | 0.29 |
PRR15 |
proline rich 15 |
752 |
0.51 |
| chr9_89952337_89952906 | 0.29 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr5_78985270_78986298 | 0.28 |
CMYA5 |
cardiomyopathy associated 5 |
84 |
0.98 |
| chr10_25007047_25007198 | 0.28 |
ARHGAP21 |
Rho GTPase activating protein 21 |
3673 |
0.34 |
| chr16_82693769_82693931 | 0.27 |
CDH13 |
cadherin 13 |
33152 |
0.22 |
| chr16_87490664_87491008 | 0.26 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
24096 |
0.15 |
| chr5_39519910_39520061 | 0.25 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
57583 |
0.16 |
| chr4_104013838_104013994 | 0.25 |
BDH2 |
3-hydroxybutyrate dehydrogenase, type 2 |
7070 |
0.22 |
| chr6_80520238_80520389 | 0.24 |
ENSG00000221470 |
. |
16289 |
0.28 |
| chr10_104530389_104530540 | 0.24 |
WBP1L |
WW domain binding protein 1-like |
5542 |
0.16 |
| chr11_116848096_116848381 | 0.24 |
SIK3 |
SIK family kinase 3 |
20478 |
0.15 |
| chr1_215256963_215257162 | 0.24 |
KCNK2 |
potassium channel, subfamily K, member 2 |
202 |
0.97 |
| chr15_89662933_89663145 | 0.23 |
ENSG00000239151 |
. |
971 |
0.55 |
| chr17_42187560_42187844 | 0.23 |
HDAC5 |
histone deacetylase 5 |
1007 |
0.34 |
| chr2_28565345_28565858 | 0.23 |
AC093690.1 |
|
32275 |
0.15 |
| chr1_7812599_7812750 | 0.23 |
CAMTA1 |
calmodulin binding transcription activator 1 |
7653 |
0.17 |
| chr2_63971408_63971559 | 0.23 |
ENSG00000221085 |
. |
48814 |
0.16 |
| chr16_80965917_80966531 | 0.23 |
CMC2 |
C-x(9)-C motif containing 2 |
66040 |
0.09 |
| chr13_60709592_60709946 | 0.22 |
DIAPH3-AS2 |
DIAPH3 antisense RNA 2 |
9063 |
0.22 |
| chr20_10536492_10536643 | 0.22 |
JAG1 |
jagged 1 |
106587 |
0.06 |
| chr12_6887362_6887730 | 0.22 |
ENSG00000244532 |
. |
5781 |
0.08 |
| chr10_33274501_33274785 | 0.22 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
6748 |
0.27 |
| chr10_17391082_17391289 | 0.22 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
22077 |
0.18 |
| chr12_7037386_7038830 | 0.22 |
ATN1 |
atrophin 1 |
632 |
0.41 |
| chr7_130589596_130590032 | 0.22 |
ENSG00000226380 |
. |
27516 |
0.22 |
| chr5_64310480_64310631 | 0.22 |
ENSG00000207439 |
. |
108641 |
0.07 |
| chr11_92117878_92118174 | 0.22 |
RP11-675M1.2 |
|
23316 |
0.23 |
| chr2_200319164_200319358 | 0.21 |
SATB2 |
SATB homeobox 2 |
1550 |
0.44 |
| chr4_46958424_46958575 | 0.21 |
GABRB1 |
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
37241 |
0.16 |
| chr8_138161471_138161805 | 0.21 |
ENSG00000206678 |
. |
44043 |
0.22 |
| chr5_160265170_160265321 | 0.21 |
ATP10B |
ATPase, class V, type 10B |
13974 |
0.28 |
| chr12_14989521_14989754 | 0.21 |
RP11-233G1.4 |
|
4389 |
0.13 |
| chr2_144709819_144710095 | 0.21 |
AC016910.1 |
|
15317 |
0.26 |
| chr9_110250307_110251795 | 0.20 |
KLF4 |
Kruppel-like factor 4 (gut) |
360 |
0.89 |
| chr4_81192543_81192855 | 0.20 |
FGF5 |
fibroblast growth factor 5 |
4906 |
0.28 |
| chr5_66564565_66564999 | 0.20 |
CD180 |
CD180 molecule |
72155 |
0.13 |
| chr2_216616741_216616892 | 0.20 |
ENSG00000212055 |
. |
126826 |
0.06 |
| chr4_81206298_81206449 | 0.20 |
FGF5 |
fibroblast growth factor 5 |
18580 |
0.23 |
| chr11_62162542_62162710 | 0.20 |
CTD-2531D15.5 |
|
1782 |
0.23 |
| chr11_57545200_57545645 | 0.20 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
3653 |
0.19 |
| chr13_37492943_37493377 | 0.20 |
SMAD9 |
SMAD family member 9 |
1215 |
0.52 |
| chr4_119950142_119950384 | 0.20 |
SYNPO2 |
synaptopodin 2 |
5637 |
0.3 |
| chr1_225837801_225838022 | 0.20 |
ENAH |
enabled homolog (Drosophila) |
2507 |
0.31 |
| chr15_91593023_91593177 | 0.19 |
AC068831.10 |
|
27248 |
0.12 |
| chr6_27647378_27647849 | 0.19 |
ENSG00000238648 |
. |
48425 |
0.11 |
| chr8_124853412_124853563 | 0.19 |
FER1L6 |
fer-1-like 6 (C. elegans) |
10740 |
0.25 |
| chr4_41614617_41614895 | 0.19 |
LIMCH1 |
LIM and calponin homology domains 1 |
30 |
0.99 |
| chr10_127104208_127104359 | 0.19 |
TEX36-AS1 |
TEX36 antisense RNA 1 |
158657 |
0.04 |
| chr3_150133757_150133908 | 0.19 |
TSC22D2 |
TSC22 domain family, member 2 |
5036 |
0.31 |
| chr9_140149846_140150685 | 0.19 |
NELFB |
negative elongation factor complex member B |
640 |
0.42 |
| chr1_29002381_29002532 | 0.19 |
GMEB1 |
glucocorticoid modulatory element binding protein 1 |
7164 |
0.14 |
| chr1_46378692_46378869 | 0.19 |
MAST2 |
microtubule associated serine/threonine kinase 2 |
480 |
0.87 |
| chr14_51862206_51862357 | 0.18 |
FRMD6 |
FERM domain containing 6 |
93574 |
0.07 |
| chr8_124775567_124775718 | 0.18 |
FAM91A1 |
family with sequence similarity 91, member A1 |
5054 |
0.24 |
| chr20_4669808_4669959 | 0.18 |
PRNP |
prion protein |
2694 |
0.3 |
| chr3_134181160_134181311 | 0.18 |
CEP63 |
centrosomal protein 63kDa |
23350 |
0.15 |
| chr9_132808161_132808319 | 0.18 |
FNBP1 |
formin binding protein 1 |
2767 |
0.27 |
| chr12_46759532_46760006 | 0.17 |
SLC38A2 |
solute carrier family 38, member 2 |
679 |
0.75 |
| chr1_17867456_17867607 | 0.17 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
1201 |
0.57 |
| chr13_45146605_45146867 | 0.17 |
TSC22D1 |
TSC22 domain family, member 1 |
3656 |
0.33 |
| chr17_57451504_57451655 | 0.17 |
ENSG00000263857 |
. |
8135 |
0.18 |
| chr17_62737056_62737243 | 0.17 |
RP13-104F24.1 |
|
17572 |
0.18 |
| chr7_97816035_97816206 | 0.17 |
BHLHA15 |
basic helix-loop-helix family, member a15 |
24619 |
0.18 |
| chr3_120215962_120216316 | 0.17 |
FSTL1 |
follistatin-like 1 |
46039 |
0.16 |
| chr11_71780711_71781020 | 0.17 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
268 |
0.8 |
| chr1_61784012_61784215 | 0.17 |
NFIA |
nuclear factor I/A |
34009 |
0.24 |
| chr2_65527534_65527956 | 0.17 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
34162 |
0.18 |
| chr2_20132866_20133200 | 0.17 |
WDR35 |
WD repeat domain 35 |
20599 |
0.18 |
| chr2_239895284_239895449 | 0.17 |
ENSG00000211566 |
. |
660 |
0.69 |
| chr8_38732199_38732350 | 0.17 |
PLEKHA2 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 |
26479 |
0.14 |
| chr13_38171648_38172094 | 0.17 |
POSTN |
periostin, osteoblast specific factor |
992 |
0.71 |
| chr4_100007531_100007682 | 0.16 |
ADH5 |
alcohol dehydrogenase 5 (class III), chi polypeptide |
1239 |
0.4 |
| chrX_107683395_107684019 | 0.16 |
COL4A5 |
collagen, type IV, alpha 5 |
595 |
0.63 |
| chr3_184037306_184037783 | 0.16 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
1 |
0.95 |
| chr17_77019963_77021139 | 0.16 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
208 |
0.91 |
| chr9_89874705_89874865 | 0.16 |
ENSG00000212421 |
. |
580 |
0.85 |
| chr19_6738037_6738395 | 0.16 |
TRIP10 |
thyroid hormone receptor interactor 10 |
280 |
0.7 |
| chr12_129339372_129339523 | 0.16 |
GLT1D1 |
glycosyltransferase 1 domain containing 1 |
1366 |
0.53 |
| chr1_150135684_150135897 | 0.16 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
6667 |
0.15 |
| chr15_73894901_73895100 | 0.16 |
NPTN |
neuroplastin |
30104 |
0.2 |
| chr20_10521227_10521501 | 0.16 |
SLX4IP |
SLX4 interacting protein |
105413 |
0.06 |
| chr11_31006065_31006277 | 0.16 |
DCDC1 |
doublecortin domain containing 1 |
8062 |
0.32 |
| chr2_190017431_190017582 | 0.16 |
ENSG00000264725 |
. |
19669 |
0.19 |
| chr11_126799438_126799589 | 0.15 |
KIRREL3-AS2 |
KIRREL3 antisense RNA 2 |
11129 |
0.21 |
| chr17_70512891_70513125 | 0.15 |
ENSG00000200783 |
. |
147283 |
0.04 |
| chr12_1153810_1153961 | 0.15 |
ERC1 |
ELKS/RAB6-interacting/CAST family member 1 |
16947 |
0.19 |
| chr2_205707475_205707729 | 0.15 |
PARD3B |
par-3 family cell polarity regulator beta |
296879 |
0.01 |
| chr19_55627735_55628805 | 0.15 |
PPP1R12C |
protein phosphatase 1, regulatory subunit 12C |
511 |
0.63 |
| chr8_12809514_12810001 | 0.15 |
KIAA1456 |
KIAA1456 |
226 |
0.95 |
| chr8_80343054_80343205 | 0.15 |
STMN2 |
stathmin-like 2 |
179920 |
0.03 |
| chr3_122201810_122202029 | 0.15 |
KPNA1 |
karyopherin alpha 1 (importin alpha 5) |
31470 |
0.14 |
| chr19_7182944_7183430 | 0.15 |
INSR |
insulin receptor |
15198 |
0.17 |
| chr18_47017567_47018883 | 0.15 |
RPL17 |
ribosomal protein L17 |
15 |
0.51 |
| chr6_138421277_138421428 | 0.15 |
PERP |
PERP, TP53 apoptosis effector |
7296 |
0.27 |
| chr9_79792511_79793707 | 0.15 |
VPS13A-AS1 |
VPS13A antisense RNA 1 |
199 |
0.85 |
| chr11_104763853_104764004 | 0.15 |
CASP12 |
caspase 12 (gene/pseudogene) |
5213 |
0.23 |
| chr16_65151579_65151789 | 0.14 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
4149 |
0.39 |
| chrX_96294671_96294822 | 0.14 |
RPA4 |
replication protein A4, 30kDa |
155839 |
0.04 |
| chr17_4090765_4090916 | 0.14 |
ANKFY1 |
ankyrin repeat and FYVE domain containing 1 |
436 |
0.68 |
| chr9_13001851_13002398 | 0.14 |
ENSG00000222658 |
. |
116804 |
0.06 |
| chr1_88292572_88292723 | 0.14 |
ENSG00000199318 |
. |
373591 |
0.01 |
| chr6_4282166_4282426 | 0.14 |
ENSG00000201185 |
. |
145901 |
0.04 |
| chr2_239791543_239791694 | 0.14 |
TWIST2 |
twist family bHLH transcription factor 2 |
34945 |
0.18 |
| chr2_192127945_192128119 | 0.14 |
MYO1B |
myosin IB |
267 |
0.95 |
| chr11_129710328_129710479 | 0.14 |
TMEM45B |
transmembrane protein 45B |
9824 |
0.26 |
| chr5_15021362_15021513 | 0.14 |
ENSG00000202269 |
. |
89592 |
0.09 |
| chr1_162534114_162534517 | 0.14 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
1512 |
0.34 |
| chr16_19124934_19125453 | 0.14 |
ITPRIPL2 |
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
61 |
0.97 |
| chr15_63525932_63526083 | 0.14 |
APH1B |
APH1B gamma secretase subunit |
42210 |
0.14 |
| chr2_190150776_190150927 | 0.14 |
ENSG00000266817 |
. |
25636 |
0.23 |
| chr6_63226419_63226570 | 0.14 |
ENSG00000216072 |
. |
166679 |
0.04 |
| chr2_192067478_192067647 | 0.14 |
MYO1B |
myosin IB |
42349 |
0.15 |
| chr4_116227889_116228040 | 0.14 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
192932 |
0.03 |
| chr16_27297585_27297841 | 0.14 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
17598 |
0.16 |
| chrX_9487811_9487962 | 0.14 |
TBL1X |
transducin (beta)-like 1X-linked |
15097 |
0.3 |
| chr12_131198863_131199152 | 0.14 |
RIMBP2 |
RIMS binding protein 2 |
1819 |
0.46 |
| chr6_117086060_117086951 | 0.14 |
FAM162B |
family with sequence similarity 162, member B |
381 |
0.87 |
| chr14_23318650_23318801 | 0.14 |
ENSG00000212335 |
. |
3212 |
0.11 |
| chr12_16501800_16502026 | 0.14 |
MGST1 |
microsomal glutathione S-transferase 1 |
1182 |
0.59 |
| chr3_189306684_189306989 | 0.14 |
TP63 |
tumor protein p63 |
42380 |
0.21 |
| chr1_42115734_42115934 | 0.14 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
50838 |
0.16 |
| chr1_36038675_36039393 | 0.13 |
TFAP2E |
transcription factor AP-2 epsilon (activating enhancer binding protein 2 epsilon) |
63 |
0.8 |
| chr6_20497460_20497742 | 0.13 |
ENSG00000201683 |
. |
2805 |
0.26 |
| chr14_37517694_37517845 | 0.13 |
RP11-537P22.2 |
|
49999 |
0.15 |
| chr16_66638822_66640143 | 0.13 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
58 |
0.95 |
| chr1_34630625_34630811 | 0.13 |
CSMD2 |
CUB and Sushi multiple domains 2 |
157 |
0.94 |
| chr8_107513799_107513950 | 0.13 |
OXR1 |
oxidation resistance 1 |
53722 |
0.17 |
| chr8_124502561_124502827 | 0.13 |
FBXO32 |
F-box protein 32 |
50752 |
0.11 |
| chr1_14081545_14081696 | 0.13 |
PRDM2 |
PR domain containing 2, with ZNF domain |
5689 |
0.27 |
| chr22_36235496_36235773 | 0.13 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
631 |
0.82 |
| chr17_64187141_64188685 | 0.13 |
CEP112 |
centrosomal protein 112kDa |
60 |
0.98 |
| chr3_147072396_147072661 | 0.13 |
RP11-649A16.1 |
|
15915 |
0.19 |
| chr9_14373044_14373195 | 0.13 |
RP11-120J1.1 |
|
25800 |
0.22 |
| chr15_61209604_61209837 | 0.13 |
RP11-554D20.1 |
|
152781 |
0.04 |
| chr16_83630209_83630360 | 0.13 |
ENSG00000263785 |
. |
88333 |
0.08 |
| chr10_18623245_18623419 | 0.13 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
6334 |
0.3 |
| chr15_70615789_70615940 | 0.13 |
ENSG00000200216 |
. |
130289 |
0.06 |
| chr7_99973393_99973544 | 0.13 |
PILRA |
paired immunoglobin-like type 2 receptor alpha |
2317 |
0.14 |
| chr15_59023939_59024090 | 0.13 |
ADAM10 |
ADAM metallopeptidase domain 10 |
17779 |
0.14 |
| chrX_51634929_51635080 | 0.13 |
MAGED1 |
melanoma antigen family D, 1 |
1625 |
0.44 |
| chr11_46311932_46312209 | 0.13 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
4607 |
0.2 |
| chr14_52708086_52708298 | 0.13 |
PTGDR |
prostaglandin D2 receptor (DP) |
26239 |
0.22 |
| chr9_77724372_77724523 | 0.13 |
OSTF1 |
osteoclast stimulating factor 1 |
20988 |
0.17 |
| chr11_131765018_131765169 | 0.13 |
AP004372.1 |
|
1909 |
0.41 |
| chr5_131793521_131793672 | 0.13 |
ENSG00000202533 |
. |
10243 |
0.13 |
| chr14_89453077_89453228 | 0.13 |
TTC8 |
tetratricopeptide repeat domain 8 |
145280 |
0.04 |
| chr6_53109316_53109571 | 0.13 |
ENSG00000206908 |
. |
25384 |
0.15 |
| chr5_33364778_33364929 | 0.13 |
CTD-2203K17.1 |
|
75872 |
0.11 |
| chr2_101435204_101435829 | 0.13 |
NPAS2 |
neuronal PAS domain protein 2 |
1098 |
0.54 |
| chr9_94184418_94184834 | 0.13 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
1518 |
0.54 |
| chr9_97151112_97151263 | 0.13 |
HIATL1 |
hippocampus abundant transcript-like 1 |
14354 |
0.22 |
| chr7_27138269_27138448 | 0.13 |
HOTAIRM1 |
HOXA transcript antisense RNA, myeloid-specific 1 |
100 |
0.91 |
| chr20_34324359_34324606 | 0.13 |
RBM39 |
RNA binding motif protein 39 |
4277 |
0.14 |
| chr5_15500949_15501183 | 0.12 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
481 |
0.89 |
| chr2_190272487_190272871 | 0.12 |
WDR75 |
WD repeat domain 75 |
33480 |
0.21 |
| chr12_120671366_120671711 | 0.12 |
PXN |
paxillin |
6891 |
0.13 |
| chrY_26417655_26417806 | 0.12 |
ENSG00000242425 |
. |
60348 |
0.14 |
| chrX_47004919_47005852 | 0.12 |
NDUFB11 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa |
482 |
0.54 |
| chr8_56805751_56805987 | 0.12 |
RP11-318K15.2 |
|
285 |
0.88 |
| chr16_88134555_88134901 | 0.12 |
BANP |
BTG3 associated nuclear protein |
131104 |
0.05 |
| chr16_59667984_59668135 | 0.12 |
ENSG00000200062 |
. |
94029 |
0.09 |
| chr18_65691781_65691932 | 0.12 |
DSEL |
dermatan sulfate epimerase-like |
507639 |
0.0 |
| chr2_24925560_24925711 | 0.12 |
ENSG00000206732 |
. |
26457 |
0.19 |
| chr15_96418443_96418594 | 0.12 |
ENSG00000222076 |
. |
129485 |
0.06 |
| chr6_16725171_16725625 | 0.12 |
RP1-151F17.1 |
|
35971 |
0.19 |
| chr12_116563914_116564400 | 0.12 |
ENSG00000207967 |
. |
22302 |
0.2 |
| chr7_22897710_22898017 | 0.12 |
ENSG00000221740 |
. |
1631 |
0.44 |
| chr9_109876837_109877058 | 0.12 |
RP11-508N12.2 |
|
11678 |
0.29 |
| chr1_214435103_214435394 | 0.12 |
SMYD2 |
SET and MYND domain containing 2 |
19328 |
0.27 |
| chr11_34653922_34654073 | 0.12 |
EHF |
ets homologous factor |
14 |
0.99 |
| chr1_203503529_203503721 | 0.12 |
OPTC |
opticin |
38527 |
0.16 |
| chr7_97952929_97953353 | 0.12 |
RP11-307C18.1 |
|
976 |
0.55 |
| chr1_161536147_161536341 | 0.12 |
RP11-5K23.5 |
|
9376 |
0.12 |
| chr12_7168097_7168259 | 0.12 |
C1S |
complement component 1, s subcomponent |
189 |
0.9 |
| chr5_10704875_10705026 | 0.12 |
CTD-2154B17.4 |
|
56227 |
0.12 |
| chr7_110752755_110752906 | 0.12 |
ENSG00000238922 |
. |
3417 |
0.32 |
| chr8_95048500_95048770 | 0.12 |
ENSG00000263855 |
. |
62553 |
0.12 |
| chr2_179314595_179315822 | 0.12 |
PRKRA |
protein kinase, interferon-inducible double stranded RNA dependent activator |
276 |
0.8 |
| chr7_52568857_52569008 | 0.12 |
ENSG00000238506 |
. |
177347 |
0.03 |
| chrX_138277340_138277570 | 0.12 |
FGF13 |
fibroblast growth factor 13 |
8232 |
0.33 |
| chr3_191888187_191888338 | 0.12 |
FGF12-AS1 |
FGF12 antisense RNA 1 |
67564 |
0.12 |
| chr18_51746912_51747063 | 0.12 |
ENSG00000207233 |
. |
1795 |
0.36 |
| chr6_56213688_56213839 | 0.12 |
RP3-445N2.1 |
|
16848 |
0.23 |
| chrX_71614971_71615122 | 0.12 |
ENSG00000222810 |
. |
18027 |
0.22 |
| chr20_33927086_33927237 | 0.12 |
FAM83C |
family with sequence similarity 83, member C |
46957 |
0.07 |
| chr5_36723248_36723399 | 0.11 |
CTD-2353F22.1 |
|
1952 |
0.47 |
| chr20_30457693_30458808 | 0.11 |
DUSP15 |
dual specificity phosphatase 15 |
125 |
0.67 |
| chr3_57051305_57051541 | 0.11 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
40020 |
0.14 |
| chr17_1986984_1987135 | 0.11 |
RP11-667K14.5 |
|
5916 |
0.11 |
| chr7_112740362_112740513 | 0.11 |
RP11-736E3.1 |
|
281 |
0.92 |
| chr3_150755555_150755706 | 0.11 |
MED12L |
mediator complex subunit 12-like |
48551 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis(GO:0060502) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0035844 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.0 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |