Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
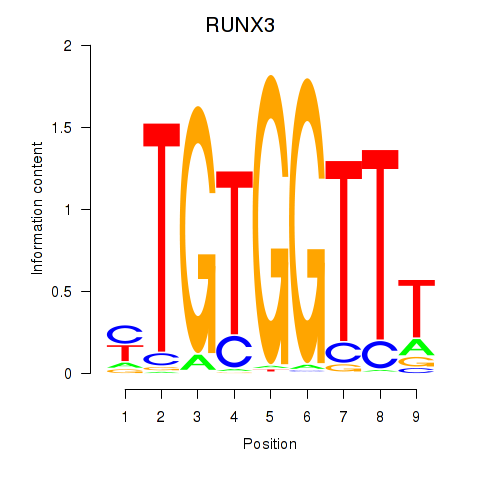
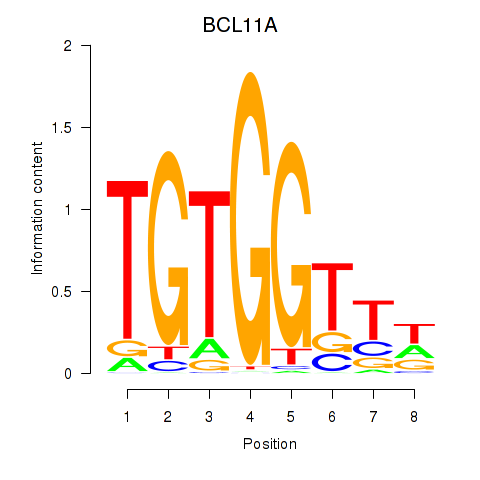
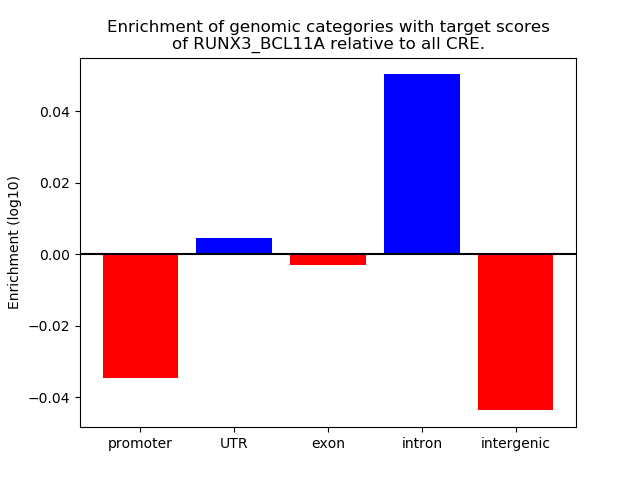
Results for RUNX3_BCL11A
Z-value: 1.28
Transcription factors associated with RUNX3_BCL11A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RUNX3
|
ENSG00000020633.14 | RUNX family transcription factor 3 |
|
BCL11A
|
ENSG00000119866.16 | BAF chromatin remodeling complex subunit BCL11A |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_60784499_60784860 | BCL11A | 3977 | 0.289631 | 0.87 | 2.1e-03 | Click! |
| chr2_60756957_60757205 | BCL11A | 23459 | 0.199334 | 0.82 | 7.0e-03 | Click! |
| chr2_60783649_60784435 | BCL11A | 3340 | 0.308308 | 0.61 | 7.8e-02 | Click! |
| chr2_60757710_60758018 | BCL11A | 22676 | 0.201763 | 0.55 | 1.3e-01 | Click! |
| chr2_60774340_60774491 | BCL11A | 6125 | 0.258308 | 0.49 | 1.8e-01 | Click! |
| chr1_25247095_25247246 | RUNX3 | 8442 | 0.232048 | -0.61 | 7.9e-02 | Click! |
| chr1_25253994_25254145 | RUNX3 | 1543 | 0.448490 | 0.60 | 8.8e-02 | Click! |
| chr1_25252382_25252533 | RUNX3 | 3155 | 0.292404 | 0.60 | 9.1e-02 | Click! |
| chr1_25255138_25255978 | RUNX3 | 54 | 0.980403 | 0.58 | 1.0e-01 | Click! |
| chr1_25252115_25252332 | RUNX3 | 3389 | 0.283851 | 0.57 | 1.1e-01 | Click! |
Activity of the RUNX3_BCL11A motif across conditions
Conditions sorted by the z-value of the RUNX3_BCL11A motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_116285976_116286984 | 0.64 |
ABLIM1 |
actin binding LIM protein 1 |
114 |
0.98 |
| chr14_91882732_91883366 | 0.50 |
CCDC88C |
coiled-coil domain containing 88C |
641 |
0.78 |
| chr12_2800154_2801438 | 0.48 |
CACNA1C-AS1 |
CACNA1C antisense RNA 1 |
430 |
0.83 |
| chrX_12974492_12974668 | 0.44 |
TMSB4X |
thymosin beta 4, X-linked |
18647 |
0.19 |
| chr1_1148809_1149188 | 0.43 |
TNFRSF4 |
tumor necrosis factor receptor superfamily, member 4 |
514 |
0.56 |
| chr21_40139305_40139870 | 0.42 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
37644 |
0.21 |
| chr19_39029395_39029671 | 0.42 |
AC067969.2 |
|
1765 |
0.24 |
| chr2_173327465_173327877 | 0.41 |
AC078883.3 |
|
3069 |
0.24 |
| chr9_116635130_116635281 | 0.39 |
ZNF618 |
zinc finger protein 618 |
3357 |
0.34 |
| chr16_85649875_85650315 | 0.39 |
GSE1 |
Gse1 coiled-coil protein |
3137 |
0.26 |
| chr2_191929470_191929872 | 0.38 |
ENSG00000231858 |
. |
43419 |
0.12 |
| chr4_84034184_84034613 | 0.36 |
PLAC8 |
placenta-specific 8 |
1470 |
0.48 |
| chr19_13215586_13215874 | 0.35 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
1755 |
0.21 |
| chr14_92301921_92302175 | 0.34 |
TC2N |
tandem C2 domains, nuclear |
736 |
0.7 |
| chr12_47609308_47609580 | 0.34 |
PCED1B |
PC-esterase domain containing 1B |
608 |
0.77 |
| chr19_16477276_16478284 | 0.34 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
5016 |
0.17 |
| chr3_39402780_39403041 | 0.34 |
SLC25A38 |
solute carrier family 25, member 38 |
21929 |
0.13 |
| chr5_39203555_39203988 | 0.32 |
FYB |
FYN binding protein |
642 |
0.81 |
| chr13_41707065_41707565 | 0.32 |
KBTBD6 |
kelch repeat and BTB (POZ) domain containing 6 |
433 |
0.79 |
| chr9_92092302_92093120 | 0.32 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
2094 |
0.37 |
| chr20_8194475_8194883 | 0.31 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
34672 |
0.21 |
| chr19_42703347_42703984 | 0.31 |
ENSG00000265122 |
. |
6435 |
0.1 |
| chr17_29640754_29641122 | 0.31 |
EVI2B |
ecotropic viral integration site 2B |
164 |
0.93 |
| chrX_57313067_57313960 | 0.31 |
FAAH2 |
fatty acid amide hydrolase 2 |
374 |
0.92 |
| chr1_117301083_117301385 | 0.30 |
CD2 |
CD2 molecule |
4145 |
0.26 |
| chrX_118815979_118816490 | 0.30 |
SEPT6 |
septin 6 |
10558 |
0.17 |
| chr16_29674660_29674967 | 0.30 |
QPRT |
quinolinate phosphoribosyltransferase |
213 |
0.51 |
| chr12_109027255_109027564 | 0.30 |
SELPLG |
selectin P ligand |
261 |
0.87 |
| chrX_70842641_70842792 | 0.30 |
ENSG00000264855 |
. |
2989 |
0.24 |
| chr1_232054576_232054868 | 0.29 |
DISC1-IT1 |
DISC1 intronic transcript 1 (non-protein coding) |
6858 |
0.32 |
| chr16_88844628_88844779 | 0.29 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
6916 |
0.09 |
| chr1_85328555_85328706 | 0.29 |
LPAR3 |
lysophosphatidic acid receptor 3 |
3212 |
0.32 |
| chr1_117298667_117299007 | 0.28 |
CD2 |
CD2 molecule |
1748 |
0.4 |
| chr2_202097823_202098101 | 0.28 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
204 |
0.94 |
| chr5_75700712_75700863 | 0.28 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
538 |
0.86 |
| chrX_129225814_129226276 | 0.28 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
18291 |
0.19 |
| chr12_10021470_10021931 | 0.28 |
CLEC2B |
C-type lectin domain family 2, member B |
1035 |
0.4 |
| chr2_54809030_54809206 | 0.27 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
23587 |
0.17 |
| chr1_209941174_209941325 | 0.27 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
584 |
0.67 |
| chr13_114549320_114549471 | 0.27 |
GAS6 |
growth arrest-specific 6 |
661 |
0.76 |
| chr14_71465889_71466177 | 0.26 |
PCNX |
pecanex homolog (Drosophila) |
13715 |
0.29 |
| chr2_158295494_158296053 | 0.26 |
CYTIP |
cytohesin 1 interacting protein |
153 |
0.96 |
| chr7_26333293_26333815 | 0.26 |
SNX10 |
sorting nexin 10 |
880 |
0.67 |
| chr20_47439188_47439680 | 0.26 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
4986 |
0.3 |
| chr9_131831774_131831925 | 0.26 |
DOLPP1 |
dolichyldiphosphatase 1 |
11530 |
0.11 |
| chr1_26644827_26645344 | 0.26 |
UBXN11 |
UBX domain protein 11 |
231 |
0.76 |
| chr22_17599001_17599302 | 0.25 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
2992 |
0.19 |
| chr8_134072568_134072719 | 0.25 |
SLA |
Src-like-adaptor |
40 |
0.98 |
| chr4_148602160_148602311 | 0.25 |
PRMT10 |
protein arginine methyltransferase 10 (putative) |
3041 |
0.27 |
| chr1_175178431_175178668 | 0.24 |
KIAA0040 |
KIAA0040 |
16470 |
0.25 |
| chr14_51288219_51288370 | 0.24 |
RP11-286O18.1 |
|
304 |
0.55 |
| chr20_49136153_49136645 | 0.24 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
9479 |
0.17 |
| chr3_113250860_113251011 | 0.24 |
SIDT1 |
SID1 transmembrane family, member 1 |
208 |
0.94 |
| chr9_78639191_78639342 | 0.24 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
71627 |
0.13 |
| chr20_33732797_33732980 | 0.24 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
71 |
0.96 |
| chr16_68306988_68307276 | 0.24 |
SLC7A6 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
250 |
0.81 |
| chr20_47441910_47442200 | 0.24 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
2365 |
0.4 |
| chr18_43914231_43915043 | 0.24 |
RNF165 |
ring finger protein 165 |
450 |
0.89 |
| chr19_41813133_41813284 | 0.24 |
CCDC97 |
coiled-coil domain containing 97 |
2886 |
0.15 |
| chr2_7866064_7866235 | 0.23 |
ENSG00000221255 |
. |
149177 |
0.05 |
| chr8_27238127_27238372 | 0.23 |
PTK2B |
protein tyrosine kinase 2 beta |
81 |
0.98 |
| chr18_67612600_67612814 | 0.23 |
CD226 |
CD226 molecule |
1948 |
0.47 |
| chr17_38018541_38018728 | 0.23 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1745 |
0.28 |
| chr15_40745675_40745826 | 0.23 |
RP11-64K12.9 |
|
1088 |
0.32 |
| chr5_118676780_118676931 | 0.23 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
7985 |
0.22 |
| chr15_85141845_85142241 | 0.23 |
ZSCAN2 |
zinc finger and SCAN domain containing 2 |
2174 |
0.19 |
| chr11_75081153_75081345 | 0.23 |
ARRB1 |
arrestin, beta 1 |
18376 |
0.12 |
| chr19_47611961_47612112 | 0.23 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
4495 |
0.17 |
| chr16_88768022_88768436 | 0.23 |
RNF166 |
ring finger protein 166 |
1218 |
0.22 |
| chr13_41579433_41579584 | 0.23 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
13942 |
0.19 |
| chr3_30653748_30654209 | 0.23 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
5885 |
0.32 |
| chr4_169680634_169680785 | 0.23 |
PALLD |
palladin, cytoskeletal associated protein |
47399 |
0.14 |
| chr13_41372571_41372761 | 0.23 |
SLC25A15 |
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
5372 |
0.18 |
| chr5_39176975_39177210 | 0.23 |
FYB |
FYN binding protein |
26037 |
0.23 |
| chr1_55678186_55678720 | 0.23 |
USP24 |
ubiquitin specific peptidase 24 |
2309 |
0.34 |
| chr1_114413652_114413958 | 0.23 |
PTPN22 |
protein tyrosine phosphatase, non-receptor type 22 (lymphoid) |
515 |
0.69 |
| chr5_133446207_133446358 | 0.23 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
4120 |
0.26 |
| chr5_40683713_40683958 | 0.23 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
4235 |
0.24 |
| chr1_36923329_36923682 | 0.23 |
MRPS15 |
mitochondrial ribosomal protein S15 |
6533 |
0.14 |
| chr5_99606023_99606303 | 0.23 |
ENSG00000207077 |
. |
116787 |
0.07 |
| chr3_150324103_150324270 | 0.23 |
SELT |
Selenoprotein T |
3063 |
0.23 |
| chr8_29954088_29954370 | 0.23 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
1064 |
0.45 |
| chr8_75201254_75201405 | 0.23 |
JPH1 |
junctophilin 1 |
32234 |
0.16 |
| chr4_154354700_154355154 | 0.22 |
KIAA0922 |
KIAA0922 |
32571 |
0.17 |
| chr12_113416849_113417235 | 0.22 |
OAS2 |
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
697 |
0.66 |
| chr17_73840046_73840332 | 0.22 |
UNC13D |
unc-13 homolog D (C. elegans) |
226 |
0.84 |
| chr15_67390777_67391884 | 0.22 |
SMAD3 |
SMAD family member 3 |
313 |
0.93 |
| chr5_156698426_156698828 | 0.22 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
2265 |
0.23 |
| chr16_68318190_68318689 | 0.22 |
ENSG00000252026 |
. |
218 |
0.84 |
| chr3_18767122_18767494 | 0.22 |
ENSG00000228956 |
. |
19928 |
0.29 |
| chr17_73064038_73064189 | 0.22 |
SLC16A5 |
solute carrier family 16 (monocarboxylate transporter), member 5 |
19709 |
0.07 |
| chr10_126428666_126428902 | 0.22 |
FAM53B |
family with sequence similarity 53, member B |
2755 |
0.24 |
| chr14_22947996_22948147 | 0.22 |
ENSG00000251002 |
. |
2207 |
0.16 |
| chr5_75839071_75839222 | 0.22 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
4088 |
0.29 |
| chr6_10529624_10529892 | 0.22 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
1169 |
0.49 |
| chrX_42793717_42793868 | 0.22 |
MAOA |
monoamine oxidase A |
721675 |
0.0 |
| chr3_142227777_142228000 | 0.22 |
RP11-383G6.3 |
|
36709 |
0.13 |
| chr6_150954320_150954515 | 0.22 |
RP11-136K14.2 |
|
8604 |
0.22 |
| chr19_35492875_35493026 | 0.21 |
GRAMD1A |
GRAM domain containing 1A |
1590 |
0.22 |
| chr12_51019673_51019824 | 0.21 |
ENSG00000207136 |
. |
7308 |
0.22 |
| chr5_171601000_171601151 | 0.21 |
STK10 |
serine/threonine kinase 10 |
14315 |
0.19 |
| chr9_128586328_128586530 | 0.21 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
41121 |
0.2 |
| chr16_50776896_50777610 | 0.21 |
RP11-327F22.1 |
|
527 |
0.39 |
| chr6_30798000_30798388 | 0.21 |
ENSG00000202241 |
. |
33833 |
0.07 |
| chr2_192058733_192058884 | 0.21 |
STAT4 |
signal transducer and activator of transcription 4 |
42486 |
0.15 |
| chr15_70851080_70851231 | 0.21 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
143465 |
0.05 |
| chr17_75663137_75663644 | 0.21 |
SEPT9 |
septin 9 |
185395 |
0.03 |
| chr20_13228054_13228205 | 0.21 |
RP5-1077I2.3 |
|
2113 |
0.33 |
| chr17_66352660_66353063 | 0.21 |
ARSG |
arylsulfatase G |
65202 |
0.09 |
| chr2_198131130_198131281 | 0.21 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
36038 |
0.14 |
| chr21_16507528_16507679 | 0.21 |
NRIP1 |
nuclear receptor interacting protein 1 |
70282 |
0.13 |
| chr14_65165620_65165918 | 0.21 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
5051 |
0.25 |
| chr14_22966536_22967037 | 0.21 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
10615 |
0.1 |
| chr8_11663519_11663931 | 0.21 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
1727 |
0.27 |
| chr18_43784563_43784787 | 0.21 |
C18orf25 |
chromosome 18 open reading frame 25 |
30675 |
0.18 |
| chr5_100067523_100067876 | 0.21 |
ENSG00000207269 |
. |
1063 |
0.67 |
| chr7_50348888_50349341 | 0.21 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
796 |
0.76 |
| chr6_135409102_135409253 | 0.20 |
HBS1L |
HBS1-like (S. cerevisiae) |
15017 |
0.21 |
| chr9_139635971_139636180 | 0.20 |
LCN10 |
lipocalin 10 |
1280 |
0.21 |
| chr13_108457167_108457318 | 0.20 |
FAM155A-IT1 |
FAM155A intronic transcript 1 (non-protein coding) |
30557 |
0.22 |
| chr15_40599321_40599893 | 0.20 |
PLCB2 |
phospholipase C, beta 2 |
419 |
0.68 |
| chr9_134603422_134603972 | 0.20 |
ENSG00000240853 |
. |
2843 |
0.28 |
| chr14_91833633_91833915 | 0.20 |
ENSG00000265856 |
. |
33717 |
0.17 |
| chr2_235397304_235397485 | 0.20 |
ARL4C |
ADP-ribosylation factor-like 4C |
7850 |
0.33 |
| chr15_75072904_75073055 | 0.20 |
CSK |
c-src tyrosine kinase |
1419 |
0.29 |
| chr18_67565113_67565264 | 0.20 |
CD226 |
CD226 molecule |
49467 |
0.18 |
| chr3_3234132_3234283 | 0.20 |
CRBN |
cereblon |
12813 |
0.2 |
| chr17_7462231_7462382 | 0.20 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
202 |
0.78 |
| chr14_102342635_102343008 | 0.20 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
7053 |
0.2 |
| chr6_108489604_108489769 | 0.20 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
281 |
0.9 |
| chr5_67551667_67551899 | 0.20 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
16057 |
0.26 |
| chr22_19792659_19792810 | 0.20 |
TBX1 |
T-box 1 |
48508 |
0.1 |
| chr19_8274834_8275489 | 0.20 |
CERS4 |
ceramide synthase 4 |
643 |
0.68 |
| chr9_7935667_7935818 | 0.20 |
TMEM261 |
transmembrane protein 261 |
135675 |
0.06 |
| chr3_15578534_15578685 | 0.20 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
15351 |
0.14 |
| chrX_144503620_144503771 | 0.20 |
SPANXN1 |
SPANX family, member N1 |
175347 |
0.03 |
| chr7_26192485_26193339 | 0.20 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
1052 |
0.53 |
| chr22_50524316_50524467 | 0.20 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
60 |
0.96 |
| chr20_31116427_31116578 | 0.20 |
C20orf112 |
chromosome 20 open reading frame 112 |
7698 |
0.18 |
| chr17_72755530_72755949 | 0.20 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
2403 |
0.16 |
| chr8_130983839_130984300 | 0.20 |
FAM49B |
family with sequence similarity 49, member B |
833 |
0.63 |
| chr21_43844485_43844636 | 0.20 |
ENSG00000252619 |
. |
6858 |
0.14 |
| chr11_3880339_3880818 | 0.19 |
STIM1 |
stromal interaction molecule 1 |
3166 |
0.13 |
| chr5_40407299_40407450 | 0.19 |
ENSG00000265615 |
. |
86954 |
0.1 |
| chr3_31481429_31481580 | 0.19 |
ENSG00000238727 |
. |
44075 |
0.19 |
| chr12_125405924_125406261 | 0.19 |
UBC |
ubiquitin C |
4178 |
0.19 |
| chr4_38511545_38511706 | 0.19 |
RP11-617D20.1 |
|
114571 |
0.06 |
| chr2_139211862_139212013 | 0.19 |
SPOPL |
speckle-type POZ protein-like |
47434 |
0.2 |
| chr10_30818638_30818892 | 0.19 |
ENSG00000239744 |
. |
26068 |
0.21 |
| chr15_64795611_64796094 | 0.19 |
ZNF609 |
zinc finger protein 609 |
4361 |
0.19 |
| chr16_50780900_50781420 | 0.19 |
CYLD |
cylindromatosis (turban tumor syndrome) |
2300 |
0.18 |
| chr6_11330937_11331205 | 0.19 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
51461 |
0.16 |
| chr2_235362717_235362976 | 0.19 |
ARL4C |
ADP-ribosylation factor-like 4C |
42398 |
0.22 |
| chr9_115918189_115918351 | 0.19 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
5048 |
0.23 |
| chr1_54751269_54751420 | 0.19 |
RP5-997D24.3 |
|
266 |
0.91 |
| chr5_134761441_134761660 | 0.19 |
C5orf20 |
chromosome 5 open reading frame 20 |
21488 |
0.12 |
| chr20_35258352_35258894 | 0.19 |
SLA2 |
Src-like-adaptor 2 |
15661 |
0.12 |
| chr7_2738603_2738843 | 0.19 |
AMZ1 |
archaelysin family metallopeptidase 1 |
10887 |
0.21 |
| chr11_67038920_67039071 | 0.19 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
5043 |
0.13 |
| chr17_75451832_75452118 | 0.19 |
SEPT9 |
septin 9 |
473 |
0.77 |
| chr6_90145242_90145393 | 0.19 |
ANKRD6 |
ankyrin repeat domain 6 |
2385 |
0.29 |
| chrX_4209642_4209793 | 0.19 |
ENSG00000264861 |
. |
291824 |
0.01 |
| chr6_22221737_22221888 | 0.19 |
RP3-404K8.2 |
|
38841 |
0.2 |
| chr16_57577463_57577729 | 0.19 |
GPR114 |
G protein-coupled receptor 114 |
995 |
0.47 |
| chr20_55975682_55975833 | 0.19 |
RP4-800J21.3 |
|
7639 |
0.16 |
| chr5_43037958_43038653 | 0.19 |
AC025171.1 |
|
3372 |
0.18 |
| chr1_150976711_150976969 | 0.19 |
FAM63A |
family with sequence similarity 63, member A |
2121 |
0.14 |
| chrX_96855091_96855242 | 0.19 |
DIAPH2-AS1 |
DIAPH2 antisense RNA 1 |
17092 |
0.31 |
| chr10_14603489_14603761 | 0.19 |
FAM107B |
family with sequence similarity 107, member B |
5446 |
0.27 |
| chr15_70550912_70551174 | 0.19 |
ENSG00000200216 |
. |
65468 |
0.14 |
| chr3_128717575_128717726 | 0.18 |
EFCC1 |
EF-hand and coiled-coil domain containing 1 |
2822 |
0.2 |
| chr10_6585169_6585583 | 0.18 |
PRKCQ |
protein kinase C, theta |
36825 |
0.22 |
| chr4_2283428_2283729 | 0.18 |
ZFYVE28 |
zinc finger, FYVE domain containing 28 |
5864 |
0.13 |
| chr12_104855264_104855415 | 0.18 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
4560 |
0.31 |
| chr2_99346083_99347252 | 0.18 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
44 |
0.98 |
| chr12_32639229_32639410 | 0.18 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
413 |
0.87 |
| chr17_65822638_65823477 | 0.18 |
BPTF |
bromodomain PHD finger transcription factor |
799 |
0.61 |
| chr16_53552293_53552836 | 0.18 |
AKTIP |
AKT interacting protein |
14241 |
0.21 |
| chr10_3917179_3917448 | 0.18 |
KLF6 |
Kruppel-like factor 6 |
89840 |
0.09 |
| chr1_19253687_19254204 | 0.18 |
IFFO2 |
intermediate filament family orphan 2 |
15459 |
0.16 |
| chr1_16470894_16471177 | 0.18 |
RP11-276H7.2 |
|
10671 |
0.12 |
| chr7_107201320_107201637 | 0.18 |
COG5 |
component of oligomeric golgi complex 5 |
2901 |
0.18 |
| chr21_36412378_36412763 | 0.18 |
RUNX1 |
runt-related transcription factor 1 |
8892 |
0.33 |
| chr4_84029715_84029866 | 0.18 |
PLAC8 |
placenta-specific 8 |
1206 |
0.55 |
| chr2_182170467_182170618 | 0.18 |
ENSG00000266705 |
. |
163 |
0.98 |
| chr10_666285_666436 | 0.18 |
RP11-809C18.3 |
|
8218 |
0.17 |
| chr7_50306566_50306717 | 0.18 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
37683 |
0.2 |
| chr2_25704888_25705039 | 0.18 |
AC104699.1 |
|
60977 |
0.13 |
| chr22_37544094_37544245 | 0.18 |
IL2RB |
interleukin 2 receptor, beta |
1861 |
0.22 |
| chr18_77161034_77161555 | 0.18 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
902 |
0.67 |
| chr20_43598945_43599305 | 0.18 |
STK4 |
serine/threonine kinase 4 |
3958 |
0.17 |
| chr5_176784624_176784924 | 0.18 |
RGS14 |
regulator of G-protein signaling 14 |
64 |
0.95 |
| chr13_51287948_51288226 | 0.17 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
93905 |
0.08 |
| chr16_31365954_31366231 | 0.17 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
363 |
0.8 |
| chr7_7960484_7960678 | 0.17 |
ENSG00000201747 |
. |
14431 |
0.18 |
| chr16_50778328_50778504 | 0.17 |
RP11-327F22.1 |
|
1690 |
0.19 |
| chr16_10940725_10940876 | 0.17 |
TVP23A |
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
28158 |
0.13 |
| chr2_44347613_44347791 | 0.17 |
ENSG00000252599 |
. |
34332 |
0.15 |
| chr7_116712743_116713170 | 0.17 |
ST7-AS2 |
ST7 antisense RNA 2 |
1822 |
0.35 |
| chr14_70161983_70162134 | 0.17 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
31559 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.1 | 0.3 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.1 | 0.2 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.3 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.1 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.2 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.0 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.2 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.2 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.2 | GO:0045911 | positive regulation of isotype switching(GO:0045830) positive regulation of DNA recombination(GO:0045911) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.4 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.2 | GO:0001787 | natural killer cell proliferation(GO:0001787) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.3 | GO:0036230 | granulocyte activation(GO:0036230) |
| 0.0 | 0.1 | GO:0002423 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.0 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.5 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0002639 | positive regulation of immunoglobulin production(GO:0002639) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0042695 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.2 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.0 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.1 | GO:0045655 | regulation of monocyte differentiation(GO:0045655) |
| 0.0 | 0.0 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.1 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.3 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.0 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0055022 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.0 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.0 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.0 | GO:0045136 | development of secondary sexual characteristics(GO:0045136) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.0 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:2000399 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 | 0.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.0 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.1 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.0 | GO:0009301 | snRNA transcription(GO:0009301) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0001914 | regulation of T cell mediated cytotoxicity(GO:0001914) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.1 | GO:1901224 | activation of NF-kappaB-inducing kinase activity(GO:0007250) NIK/NF-kappaB signaling(GO:0038061) regulation of NIK/NF-kappaB signaling(GO:1901222) positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:1901862 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.0 | 0.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0002836 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.0 | GO:0002709 | regulation of T cell mediated immunity(GO:0002709) |
| 0.0 | 0.0 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.0 | GO:0043501 | skeletal muscle adaptation(GO:0043501) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:2001021 | negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.0 | 0.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.0 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0038201 | TORC1 complex(GO:0031931) TOR complex(GO:0038201) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.5 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.1 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.3 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0043499 | obsolete eukaryotic cell surface binding(GO:0043499) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.4 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0046978 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 1.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.6 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.3 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.0 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.1 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |