Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
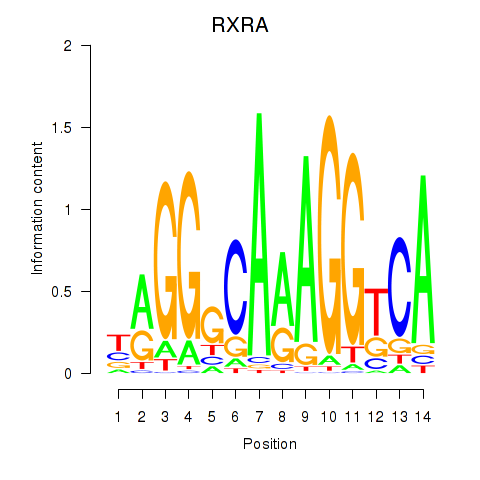
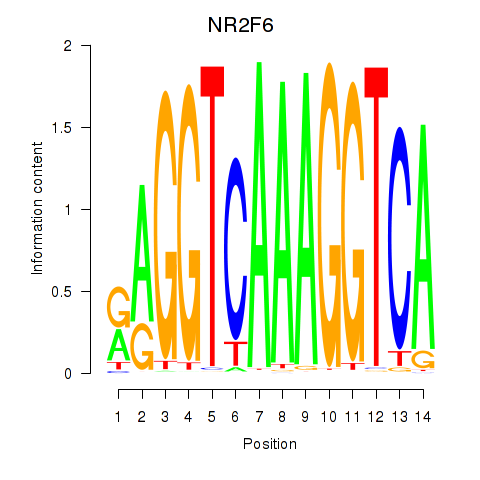
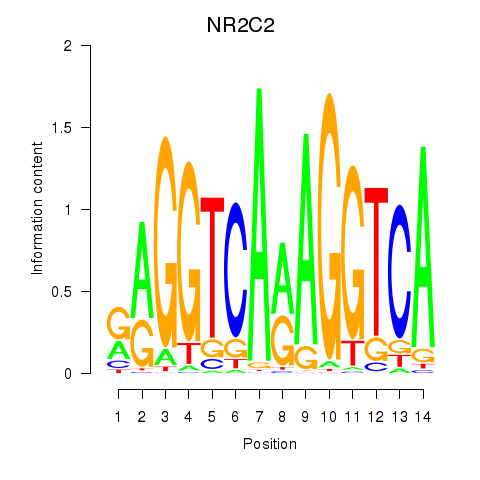
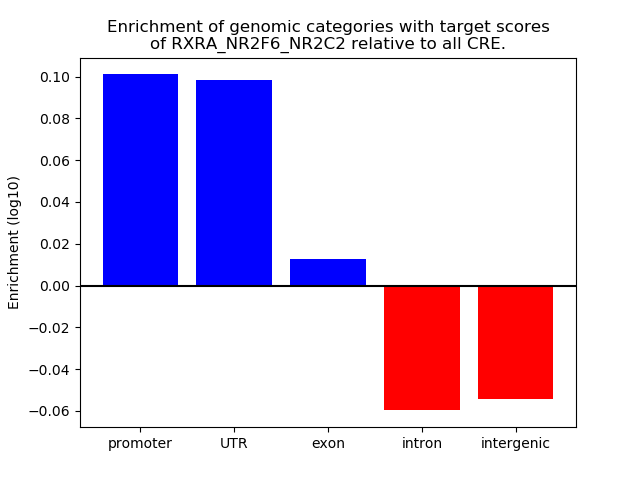
Results for RXRA_NR2F6_NR2C2
Z-value: 1.10
Transcription factors associated with RXRA_NR2F6_NR2C2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRA
|
ENSG00000186350.8 | retinoid X receptor alpha |
|
NR2F6
|
ENSG00000160113.5 | nuclear receptor subfamily 2 group F member 6 |
|
NR2C2
|
ENSG00000177463.11 | nuclear receptor subfamily 2 group C member 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_15030879_15031030 | NR2C2 | 14468 | 0.201569 | 0.64 | 6.1e-02 | Click! |
| chr3_15001001_15001179 | NR2C2 | 2824 | 0.283320 | -0.48 | 1.9e-01 | Click! |
| chr3_15052457_15052608 | NR2C2 | 7110 | 0.219334 | -0.22 | 5.8e-01 | Click! |
| chr3_15030493_15030644 | NR2C2 | 14854 | 0.200687 | 0.17 | 6.5e-01 | Click! |
| chr3_15028082_15028233 | NR2C2 | 17265 | 0.195105 | 0.13 | 7.5e-01 | Click! |
| chr19_17356354_17356505 | NR2F6 | 320 | 0.738809 | 0.33 | 3.8e-01 | Click! |
| chr19_17355272_17355498 | NR2F6 | 72 | 0.926116 | -0.32 | 4.0e-01 | Click! |
| chr19_17355503_17355912 | NR2F6 | 394 | 0.668365 | -0.20 | 6.1e-01 | Click! |
| chr19_17357023_17357252 | NR2F6 | 388 | 0.672975 | -0.10 | 8.1e-01 | Click! |
| chr19_17355975_17356268 | NR2F6 | 628 | 0.474279 | -0.06 | 8.8e-01 | Click! |
| chr9_137296411_137296683 | RXRA | 1881 | 0.410203 | 0.85 | 3.7e-03 | Click! |
| chr9_137326628_137326779 | RXRA | 28275 | 0.207889 | -0.82 | 6.6e-03 | Click! |
| chr9_137160017_137160168 | RXRA | 58334 | 0.129573 | 0.78 | 1.3e-02 | Click! |
| chr9_137335451_137335655 | RXRA | 37125 | 0.185535 | 0.74 | 2.1e-02 | Click! |
| chr9_137297323_137297474 | RXRA | 1030 | 0.613547 | 0.72 | 3.0e-02 | Click! |
Activity of the RXRA_NR2F6_NR2C2 motif across conditions
Conditions sorted by the z-value of the RXRA_NR2F6_NR2C2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
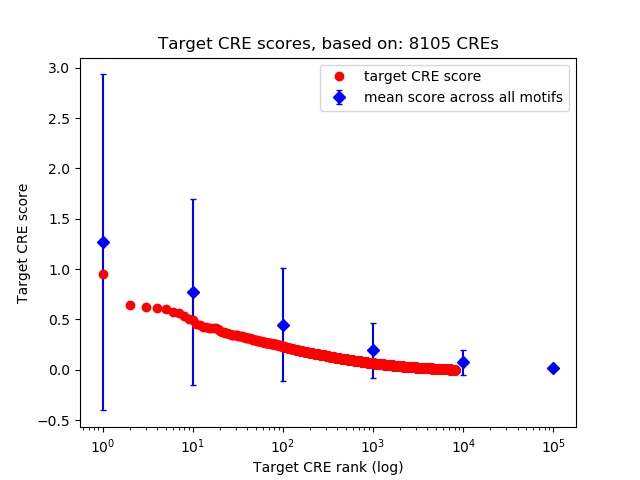
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_10416066_10416280 | 0.96 |
ZGLP1 |
zinc finger, GATA-like protein 1 |
4286 |
0.08 |
| chr17_79879240_79879391 | 0.65 |
SIRT7 |
sirtuin 7 |
116 |
0.85 |
| chr6_14721018_14721200 | 0.62 |
ENSG00000206960 |
. |
74343 |
0.13 |
| chrX_65235295_65235677 | 0.61 |
ENSG00000207939 |
. |
3226 |
0.26 |
| chr12_54694994_54695195 | 0.60 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
108 |
0.6 |
| chr12_1922066_1922217 | 0.58 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
1184 |
0.5 |
| chr22_37260110_37260261 | 0.56 |
NCF4 |
neutrophil cytosolic factor 4, 40kDa |
3155 |
0.18 |
| chr14_73931523_73931791 | 0.54 |
ENSG00000251393 |
. |
2528 |
0.22 |
| chr10_129678816_129678967 | 0.50 |
CLRN3 |
clarin 3 |
12320 |
0.22 |
| chr16_47050823_47051091 | 0.49 |
RP11-169E6.1 |
|
43057 |
0.14 |
| chr9_70850762_70850954 | 0.45 |
CBWD3 |
COBW domain containing 3 |
5539 |
0.19 |
| chr7_143078673_143079796 | 0.45 |
ZYX |
zyxin |
234 |
0.85 |
| chr11_1781045_1781196 | 0.43 |
AC068580.6 |
|
458 |
0.52 |
| chr6_109561771_109561922 | 0.42 |
ENSG00000238474 |
. |
50715 |
0.12 |
| chr19_18334931_18335362 | 0.42 |
PDE4C |
phosphodiesterase 4C, cAMP-specific |
157 |
0.91 |
| chr6_13274566_13274804 | 0.42 |
RP1-257A7.4 |
|
394 |
0.82 |
| chr17_81011742_81011893 | 0.41 |
B3GNTL1 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
2131 |
0.35 |
| chr2_60783649_60784435 | 0.41 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
3340 |
0.31 |
| chr20_4807814_4807965 | 0.40 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
3598 |
0.24 |
| chr4_6927779_6927930 | 0.39 |
TBC1D14 |
TBC1 domain family, member 14 |
15879 |
0.16 |
| chr3_171684620_171684771 | 0.37 |
TMEM212-IT1 |
TMEM212 intronic transcript 1 (non-protein coding) |
72469 |
0.1 |
| chr11_72448382_72448649 | 0.37 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
10342 |
0.12 |
| chr15_77711149_77711991 | 0.37 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
872 |
0.51 |
| chr9_35685699_35685887 | 0.36 |
TPM2 |
tropomyosin 2 (beta) |
330 |
0.73 |
| chr6_31927037_31927267 | 0.35 |
SKIV2L |
superkiller viralicidic activity 2-like (S. cerevisiae) |
204 |
0.52 |
| chr9_69268507_69268733 | 0.35 |
CBWD6 |
COBW domain containing 6 |
6027 |
0.21 |
| chr18_2049997_2050208 | 0.35 |
METTL4 |
methyltransferase like 4 |
504589 |
0.0 |
| chr1_153362333_153362484 | 0.35 |
S100A8 |
S100 calcium binding protein A8 |
1044 |
0.37 |
| chr14_20896169_20896332 | 0.34 |
KLHL33 |
kelch-like family member 33 |
7551 |
0.08 |
| chr11_113954202_113954403 | 0.34 |
ENSG00000221112 |
. |
3349 |
0.28 |
| chr19_47234689_47234868 | 0.34 |
CTB-174O21.2 |
|
2918 |
0.12 |
| chr5_36067012_36067195 | 0.34 |
UGT3A2 |
UDP glycosyltransferase 3 family, polypeptide A2 |
110 |
0.97 |
| chr17_7123160_7124223 | 0.34 |
ACADVL |
acyl-CoA dehydrogenase, very long chain |
424 |
0.48 |
| chr11_22688489_22688994 | 0.33 |
GAS2 |
growth arrest-specific 2 |
119 |
0.97 |
| chr12_56537034_56537185 | 0.33 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
8931 |
0.06 |
| chr2_219646650_219647418 | 0.33 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
555 |
0.7 |
| chr16_1990929_1991093 | 0.32 |
MSRB1 |
methionine sulfoxide reductase B1 |
2113 |
0.1 |
| chr22_32033546_32033968 | 0.32 |
PISD |
phosphatidylserine decarboxylase |
634 |
0.74 |
| chr20_4943157_4943308 | 0.32 |
SLC23A2 |
solute carrier family 23 (ascorbic acid transporter), member 2 |
38913 |
0.14 |
| chr1_33463602_33463869 | 0.32 |
RP1-117O3.2 |
|
11059 |
0.14 |
| chr2_113592071_113592291 | 0.31 |
IL1B |
interleukin 1, beta |
1830 |
0.31 |
| chr2_234257769_234257933 | 0.31 |
RP11-400N9.1 |
|
4211 |
0.17 |
| chr7_41731347_41731498 | 0.31 |
INHBA-AS1 |
INHBA antisense RNA 1 |
2092 |
0.34 |
| chr8_38722630_38723350 | 0.30 |
RP11-723D22.3 |
|
19033 |
0.16 |
| chr8_68880458_68880609 | 0.30 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
16180 |
0.28 |
| chr15_49087139_49087290 | 0.30 |
RP11-485O10.2 |
|
11827 |
0.17 |
| chr1_226374129_226374763 | 0.30 |
ACBD3 |
acyl-CoA binding domain containing 3 |
15 |
0.98 |
| chr20_62713551_62713939 | 0.30 |
C20orf201 |
chromosome 20 open reading frame 201 |
1967 |
0.15 |
| chr1_12079558_12080233 | 0.30 |
MIIP |
migration and invasion inhibitory protein |
305 |
0.84 |
| chr12_96514891_96515042 | 0.30 |
ENSG00000266889 |
. |
18939 |
0.19 |
| chr19_10362934_10363426 | 0.30 |
MRPL4 |
mitochondrial ribosomal protein L4 |
228 |
0.83 |
| chr2_232545912_232546437 | 0.29 |
MGC4771 |
|
25447 |
0.14 |
| chr1_2987231_2987751 | 0.29 |
PRDM16 |
PR domain containing 16 |
1716 |
0.34 |
| chr19_6738037_6738395 | 0.29 |
TRIP10 |
thyroid hormone receptor interactor 10 |
280 |
0.7 |
| chr10_73511646_73511797 | 0.28 |
C10orf54 |
chromosome 10 open reading frame 54 |
5666 |
0.21 |
| chr1_235091297_235091455 | 0.28 |
ENSG00000239690 |
. |
51443 |
0.16 |
| chr21_39756340_39756646 | 0.28 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
87645 |
0.09 |
| chr17_48367766_48367999 | 0.28 |
RP11-893F2.9 |
|
2666 |
0.19 |
| chrX_117858717_117858999 | 0.28 |
IL13RA1 |
interleukin 13 receptor, alpha 1 |
2677 |
0.32 |
| chr14_60620645_60620999 | 0.28 |
DHRS7 |
dehydrogenase/reductase (SDR family) member 7 |
930 |
0.61 |
| chr15_53495921_53496072 | 0.27 |
ONECUT1 |
one cut homeobox 1 |
413787 |
0.01 |
| chrX_2809831_2809982 | 0.27 |
ARSD-AS1 |
ARSD antisense RNA 1 |
13039 |
0.15 |
| chr16_28506110_28506360 | 0.27 |
APOBR |
apolipoprotein B receptor |
242 |
0.75 |
| chr11_72527375_72527681 | 0.27 |
ATG16L2 |
autophagy related 16-like 2 (S. cerevisiae) |
2051 |
0.23 |
| chr12_54688592_54688942 | 0.27 |
NFE2 |
nuclear factor, erythroid 2 |
775 |
0.4 |
| chr22_51020587_51021277 | 0.27 |
CHKB |
choline kinase beta |
496 |
0.39 |
| chr20_1876393_1876635 | 0.27 |
SIRPA |
signal-regulatory protein alpha |
572 |
0.81 |
| chr20_48808829_48809749 | 0.26 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
1913 |
0.35 |
| chr6_149450767_149450918 | 0.26 |
ENSG00000263481 |
. |
47088 |
0.15 |
| chr16_85325294_85325445 | 0.26 |
ENSG00000266307 |
. |
14562 |
0.24 |
| chr13_114271046_114271197 | 0.26 |
TFDP1 |
transcription factor Dp-1 |
31380 |
0.15 |
| chr19_43849967_43850118 | 0.26 |
CD177 |
CD177 molecule |
7783 |
0.17 |
| chr8_33412041_33412395 | 0.26 |
RNF122 |
ring finger protein 122 |
12425 |
0.13 |
| chr7_106065721_106066226 | 0.26 |
CTB-111H14.1 |
|
79312 |
0.1 |
| chr11_6656944_6657095 | 0.26 |
RP11-732A19.6 |
|
5185 |
0.09 |
| chr19_48866469_48867161 | 0.26 |
TMEM143 |
transmembrane protein 143 |
116 |
0.89 |
| chr19_36399282_36399433 | 0.26 |
TYROBP |
TYRO protein tyrosine kinase binding protein |
160 |
0.9 |
| chr12_109122720_109122871 | 0.26 |
CORO1C |
coronin, actin binding protein, 1C |
1532 |
0.34 |
| chr15_70384544_70384773 | 0.26 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
2466 |
0.34 |
| chr3_64811829_64811980 | 0.25 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
138228 |
0.05 |
| chr17_48205902_48206073 | 0.25 |
SAMD14 |
sterile alpha motif domain containing 14 |
1131 |
0.31 |
| chr17_4615541_4615774 | 0.25 |
ARRB2 |
arrestin, beta 2 |
1663 |
0.18 |
| chr2_113945024_113945175 | 0.25 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
8758 |
0.14 |
| chr17_29701117_29701268 | 0.25 |
NF1 |
neurofibromin 1 |
15626 |
0.14 |
| chr15_99417963_99418437 | 0.25 |
IGF1R |
insulin-like growth factor 1 receptor |
15370 |
0.2 |
| chr2_7075634_7075785 | 0.25 |
RNF144A |
ring finger protein 144A |
2535 |
0.29 |
| chr1_160832331_160832502 | 0.25 |
CD244 |
CD244 molecule, natural killer cell receptor 2B4 |
74 |
0.97 |
| chr19_43891664_43891815 | 0.25 |
TEX101 |
testis expressed 101 |
13909 |
0.15 |
| chr18_2984866_2985017 | 0.24 |
LPIN2 |
lipin 2 |
2070 |
0.27 |
| chr10_73456429_73456580 | 0.24 |
C10orf105 |
chromosome 10 open reading frame 105 |
23074 |
0.2 |
| chr10_14730758_14730909 | 0.24 |
ENSG00000201766 |
. |
25340 |
0.19 |
| chr6_56668655_56668840 | 0.24 |
DST |
dystonin |
18070 |
0.23 |
| chr7_95238082_95238332 | 0.24 |
AC002451.3 |
|
901 |
0.62 |
| chr1_236056109_236056279 | 0.24 |
LYST |
lysosomal trafficking regulator |
9322 |
0.17 |
| chr7_139530944_139531095 | 0.24 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
1909 |
0.44 |
| chr12_129338589_129338740 | 0.24 |
GLT1D1 |
glycosyltransferase 1 domain containing 1 |
583 |
0.82 |
| chr20_32399297_32399983 | 0.24 |
CHMP4B |
charged multivesicular body protein 4B |
530 |
0.55 |
| chr12_111843881_111845308 | 0.23 |
SH2B3 |
SH2B adaptor protein 3 |
842 |
0.62 |
| chr20_17660855_17661298 | 0.23 |
RRBP1 |
ribosome binding protein 1 |
1629 |
0.4 |
| chr11_67043892_67044043 | 0.23 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
10015 |
0.11 |
| chr5_77306460_77306689 | 0.23 |
AP3B1 |
adaptor-related protein complex 3, beta 1 subunit |
28400 |
0.21 |
| chr1_147119514_147119665 | 0.23 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
11527 |
0.22 |
| chr5_149465296_149465474 | 0.23 |
CSF1R |
colony stimulating factor 1 receptor |
605 |
0.69 |
| chr10_30831582_30831733 | 0.23 |
ENSG00000239744 |
. |
13176 |
0.24 |
| chrX_19904584_19904836 | 0.23 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
867 |
0.7 |
| chr16_66638822_66640143 | 0.23 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
58 |
0.95 |
| chrX_153659493_153659733 | 0.22 |
ATP6AP1 |
ATPase, H+ transporting, lysosomal accessory protein 1 |
2604 |
0.11 |
| chr19_1154555_1155129 | 0.22 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
293 |
0.85 |
| chr7_105317322_105317473 | 0.22 |
ATXN7L1 |
ataxin 7-like 1 |
2212 |
0.41 |
| chr7_99954721_99954872 | 0.22 |
PILRB |
paired immunoglobin-like type 2 receptor beta |
207 |
0.86 |
| chr21_44158180_44158391 | 0.22 |
AP001627.1 |
|
3583 |
0.28 |
| chr8_142105945_142106235 | 0.22 |
DENND3 |
DENN/MADD domain containing 3 |
21287 |
0.19 |
| chr15_74232964_74233156 | 0.22 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
12471 |
0.14 |
| chr11_47416116_47416267 | 0.22 |
RP11-750H9.5 |
|
1000 |
0.33 |
| chr11_75474128_75474279 | 0.22 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
3646 |
0.13 |
| chr19_1068246_1068401 | 0.22 |
HMHA1 |
histocompatibility (minor) HA-1 |
826 |
0.4 |
| chr6_76317008_76317159 | 0.22 |
SENP6 |
SUMO1/sentrin specific peptidase 6 |
5320 |
0.19 |
| chr19_676350_676550 | 0.21 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
58 |
0.95 |
| chr20_31061401_31061552 | 0.21 |
C20orf112 |
chromosome 20 open reading frame 112 |
9798 |
0.18 |
| chr7_40591526_40591750 | 0.21 |
AC004988.1 |
|
5111 |
0.35 |
| chr9_35730052_35730203 | 0.21 |
CREB3 |
cAMP responsive element binding protein 3 |
2205 |
0.12 |
| chr12_562387_562595 | 0.21 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
7039 |
0.21 |
| chr1_23850782_23850933 | 0.21 |
E2F2 |
E2F transcription factor 2 |
6855 |
0.18 |
| chr18_9827953_9828104 | 0.21 |
ENSG00000242651 |
. |
2596 |
0.28 |
| chr9_110253387_110253828 | 0.21 |
KLF4 |
Kruppel-like factor 4 (gut) |
844 |
0.66 |
| chr11_121325996_121326147 | 0.21 |
RP11-730K11.1 |
|
2349 |
0.33 |
| chr6_110118234_110118385 | 0.21 |
FIG4 |
FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) |
11100 |
0.28 |
| chr9_184980_185177 | 0.21 |
CBWD1 |
COBW domain containing 1 |
6006 |
0.2 |
| chr20_36824815_36825080 | 0.20 |
TGM2 |
transglutaminase 2 |
29967 |
0.14 |
| chr10_98418237_98418708 | 0.20 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
10896 |
0.21 |
| chr22_31625332_31625483 | 0.20 |
ENSG00000202019 |
. |
751 |
0.5 |
| chr10_29977190_29977386 | 0.20 |
ENSG00000222092 |
. |
23556 |
0.21 |
| chr9_123500093_123500244 | 0.20 |
MEGF9 |
multiple EGF-like-domains 9 |
23420 |
0.2 |
| chr17_25859259_25859414 | 0.20 |
KSR1 |
kinase suppressor of ras 1 |
50611 |
0.13 |
| chr6_131895362_131895513 | 0.20 |
ARG1 |
arginase 1 |
1072 |
0.55 |
| chr16_57126935_57127447 | 0.20 |
CPNE2 |
copine II |
672 |
0.49 |
| chr15_58438968_58439119 | 0.20 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
8019 |
0.23 |
| chr2_85727105_85727457 | 0.20 |
ENSG00000266577 |
. |
32468 |
0.08 |
| chr19_6067059_6067210 | 0.20 |
CTC-232P5.3 |
|
830 |
0.55 |
| chr11_47416652_47416817 | 0.20 |
RP11-750H9.5 |
|
457 |
0.65 |
| chr1_12046473_12046624 | 0.20 |
MFN2 |
mitofusin 2 |
4499 |
0.15 |
| chr22_36795299_36795450 | 0.20 |
MYH9 |
myosin, heavy chain 9, non-muscle |
11311 |
0.17 |
| chr16_20756487_20756638 | 0.20 |
THUMPD1 |
THUMP domain containing 1 |
3156 |
0.18 |
| chr17_36584744_36585461 | 0.20 |
ARHGAP23 |
Rho GTPase activating protein 23 |
372 |
0.84 |
| chr3_53201341_53201492 | 0.19 |
PRKCD |
protein kinase C, delta |
2271 |
0.28 |
| chr16_90033999_90034150 | 0.19 |
RP11-566K11.5 |
|
2125 |
0.18 |
| chr2_102609682_102609833 | 0.19 |
IL1R2 |
interleukin 1 receptor, type II |
1451 |
0.51 |
| chr7_76033306_76033511 | 0.19 |
SRCRB4D |
scavenger receptor cysteine rich domain containing, group B (4 domains) |
5604 |
0.17 |
| chr6_33172447_33173020 | 0.19 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
314 |
0.73 |
| chr6_49603951_49604102 | 0.19 |
RHAG |
Rh-associated glycoprotein |
526 |
0.83 |
| chr2_27602964_27603512 | 0.19 |
ZNF513 |
zinc finger protein 513 |
73 |
0.93 |
| chr1_169861896_169862644 | 0.19 |
SCYL3 |
SCY1-like 3 (S. cerevisiae) |
806 |
0.65 |
| chr13_108974971_108975122 | 0.19 |
ENSG00000223177 |
. |
21367 |
0.23 |
| chr12_89551363_89551514 | 0.19 |
ENSG00000238302 |
. |
124624 |
0.06 |
| chr17_73305250_73305401 | 0.19 |
GRB2 |
growth factor receptor-bound protein 2 |
12509 |
0.09 |
| chr1_174938035_174938186 | 0.19 |
RABGAP1L |
RAB GTPase activating protein 1-like |
4205 |
0.17 |
| chr5_101631358_101631636 | 0.19 |
SLCO4C1 |
solute carrier organic anion transporter family, member 4C1 |
756 |
0.68 |
| chr2_70172880_70173031 | 0.19 |
ENSG00000239072 |
. |
9822 |
0.12 |
| chr2_113190375_113190911 | 0.19 |
RGPD8 |
RANBP2-like and GRIP domain containing 8 |
453 |
0.86 |
| chr19_12899418_12899647 | 0.19 |
ENSG00000263800 |
. |
1590 |
0.15 |
| chr19_42133062_42133277 | 0.19 |
CEACAM4 |
carcinoembryonic antigen-related cell adhesion molecule 4 |
273 |
0.89 |
| chr12_105122483_105122634 | 0.19 |
ENSG00000222579 |
. |
123044 |
0.05 |
| chr9_117895091_117895381 | 0.18 |
DEC1 |
deleted in esophageal cancer 1 |
8861 |
0.25 |
| chr4_353397_353548 | 0.18 |
ENSG00000207642 |
. |
9526 |
0.17 |
| chr2_61153613_61153764 | 0.18 |
ENSG00000201076 |
. |
15250 |
0.18 |
| chr1_156474559_156474783 | 0.18 |
MEF2D |
myocyte enhancer factor 2D |
4051 |
0.14 |
| chr7_642417_642568 | 0.18 |
PRKAR1B |
protein kinase, cAMP-dependent, regulatory, type I, beta |
172 |
0.69 |
| chr17_77005019_77005688 | 0.18 |
CANT1 |
calcium activated nucleotidase 1 |
459 |
0.76 |
| chr16_89042423_89042673 | 0.18 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
853 |
0.55 |
| chr11_85781008_85781159 | 0.18 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
159 |
0.96 |
| chr3_37284874_37285642 | 0.18 |
RP11-259K5.2 |
|
410 |
0.48 |
| chr22_37257050_37257437 | 0.18 |
NCF4 |
neutrophil cytosolic factor 4, 40kDa |
213 |
0.91 |
| chr2_136690107_136690258 | 0.18 |
DARS |
aspartyl-tRNA synthetase |
12059 |
0.21 |
| chr1_33812503_33812654 | 0.18 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
2908 |
0.16 |
| chr1_154989875_154990097 | 0.18 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
3062 |
0.1 |
| chr9_34179429_34180002 | 0.18 |
UBAP1 |
ubiquitin associated protein 1 |
679 |
0.62 |
| chr1_10475436_10475736 | 0.18 |
APITD1 |
apoptosis-inducing, TAF9-like domain 1 |
14573 |
0.1 |
| chr1_155202832_155202983 | 0.18 |
ENSG00000216109 |
. |
3721 |
0.07 |
| chr15_57071498_57071649 | 0.18 |
ZNF280D |
zinc finger protein 280D |
45289 |
0.19 |
| chr10_21814018_21814991 | 0.18 |
SKIDA1 |
SKI/DACH domain containing 1 |
107 |
0.95 |
| chr1_99729766_99730318 | 0.18 |
LPPR4 |
Lipid phosphate phosphatase-related protein type 4 |
194 |
0.97 |
| chrX_118894160_118894311 | 0.18 |
SOWAHD |
sosondowah ankyrin repeat domain family member D |
1659 |
0.32 |
| chr1_38467251_38467531 | 0.18 |
FHL3 |
four and a half LIM domains 3 |
3786 |
0.15 |
| chr15_40625954_40626153 | 0.18 |
ENSG00000252714 |
. |
2204 |
0.13 |
| chr2_45170446_45170597 | 0.18 |
SIX3-AS1 |
SIX3 antisense RNA 1 |
1509 |
0.36 |
| chrX_138285953_138286466 | 0.17 |
FGF13 |
fibroblast growth factor 13 |
60 |
0.99 |
| chr2_231735442_231735593 | 0.17 |
ITM2C |
integral membrane protein 2C |
5138 |
0.19 |
| chr9_139928371_139928559 | 0.17 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
1003 |
0.23 |
| chr9_134460250_134460544 | 0.17 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
36828 |
0.13 |
| chr2_11828981_11829132 | 0.17 |
AC106875.1 |
|
7469 |
0.13 |
| chr5_124083658_124084316 | 0.17 |
ZNF608 |
zinc finger protein 608 |
513 |
0.71 |
| chr9_35489468_35490886 | 0.17 |
RUSC2 |
RUN and SH3 domain containing 2 |
53 |
0.97 |
| chr1_92941939_92942090 | 0.17 |
GFI1 |
growth factor independent 1 transcription repressor |
7497 |
0.27 |
| chr20_30125745_30125896 | 0.17 |
HM13 |
histocompatibility (minor) 13 |
23496 |
0.1 |
| chr19_16482178_16482409 | 0.17 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
9529 |
0.15 |
| chr17_75382520_75382671 | 0.17 |
SEPT9 |
septin 9 |
6390 |
0.17 |
| chr19_1019836_1021109 | 0.17 |
TMEM259 |
transmembrane protein 259 |
373 |
0.62 |
| chr3_10231486_10231637 | 0.17 |
IRAK2 |
interleukin-1 receptor-associated kinase 2 |
25012 |
0.1 |
| chrX_118894330_118894481 | 0.17 |
SOWAHD |
sosondowah ankyrin repeat domain family member D |
1829 |
0.3 |
| chr17_1090163_1090534 | 0.17 |
ABR |
active BCR-related |
268 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.4 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.1 | 0.2 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.1 | 0.4 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.2 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.2 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.4 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0015851 | nucleobase transport(GO:0015851) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.0 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.2 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.0 | 0.2 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.0 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.1 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:0043462 | regulation of ATPase activity(GO:0043462) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.1 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0045217 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.2 | GO:0033003 | regulation of mast cell activation(GO:0033003) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.4 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0033605 | positive regulation of catecholamine secretion(GO:0033605) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.0 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.0 | GO:0045349 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.0 | GO:0003160 | endocardium development(GO:0003157) endocardium morphogenesis(GO:0003160) endocardium formation(GO:0060214) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.5 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0038201 | TOR complex(GO:0038201) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.1 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.6 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.2 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0032129 | NAD-dependent histone deacetylase activity(GO:0017136) histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H3-K9 specific)(GO:0032129) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent protein deacetylase activity(GO:0034979) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.0 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.0 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.0 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |