Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
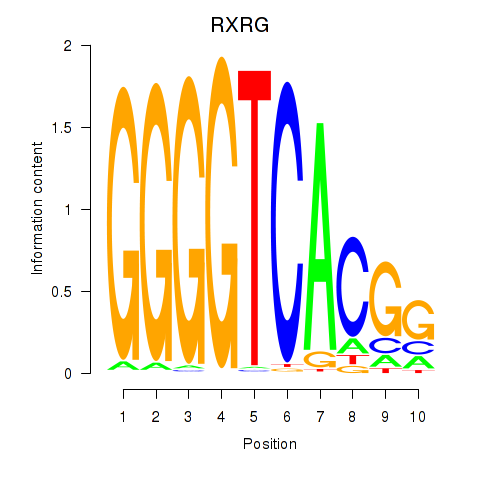
Results for RXRG
Z-value: 0.95
Transcription factors associated with RXRG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
RXRG
|
ENSG00000143171.8 | retinoid X receptor gamma |
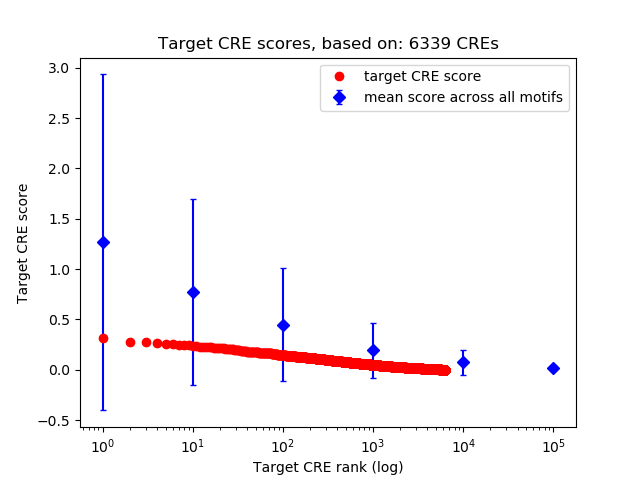
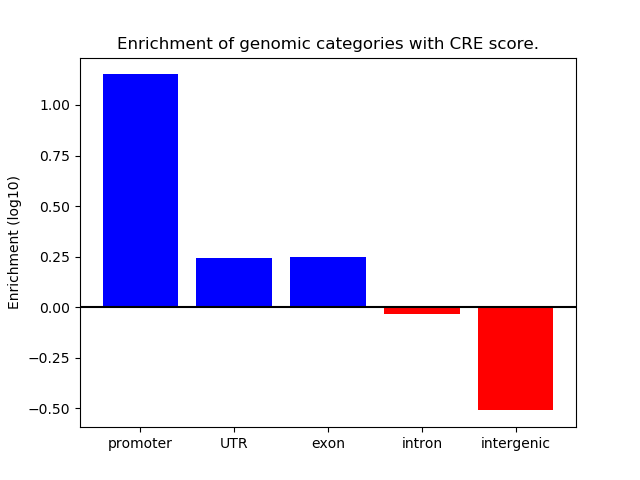
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_165394173_165394324 | RXRG | 20185 | 0.213009 | 0.54 | 1.4e-01 | Click! |
| chr1_165393775_165394037 | RXRG | 20527 | 0.212169 | 0.43 | 2.5e-01 | Click! |
| chr1_165393515_165393674 | RXRG | 20839 | 0.211397 | 0.29 | 4.5e-01 | Click! |
| chr1_165414435_165414651 | RXRG | 110 | 0.975187 | 0.28 | 4.6e-01 | Click! |
| chr1_165414145_165414329 | RXRG | 196 | 0.957396 | 0.13 | 7.3e-01 | Click! |
Activity of the RXRG motif across conditions
Conditions sorted by the z-value of the RXRG motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
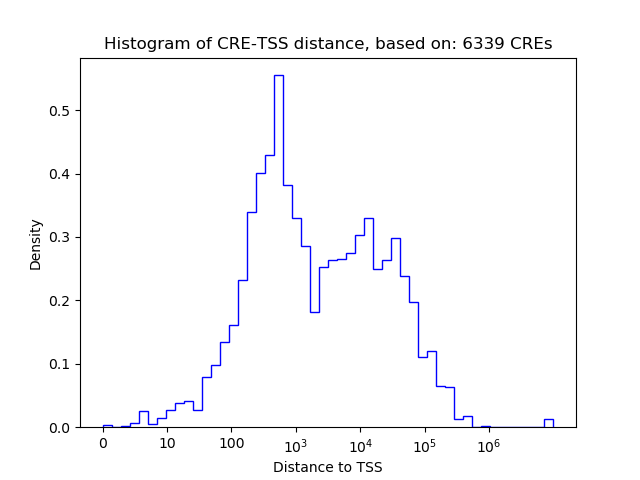
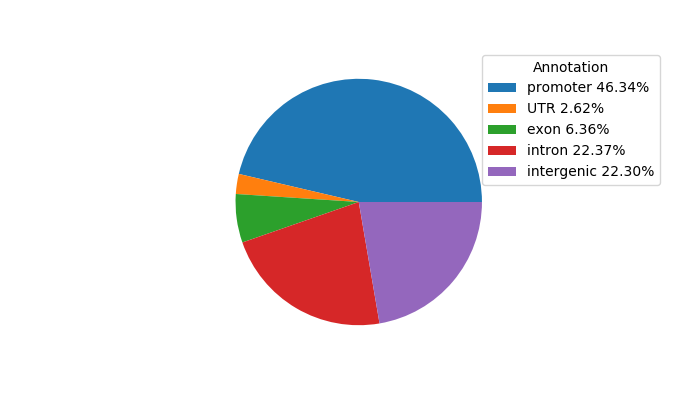
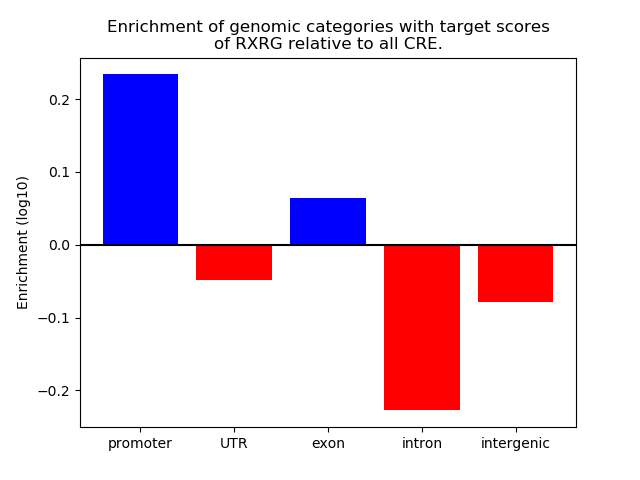
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_3749707_3750198 | 0.32 |
RP11-420L9.5 |
|
1393 |
0.41 |
| chr2_243030793_243031816 | 0.28 |
AC093642.5 |
|
460 |
0.62 |
| chr3_50193036_50193699 | 0.28 |
RP11-493K19.3 |
|
86 |
0.87 |
| chr3_184280128_184280591 | 0.27 |
EPHB3 |
EPH receptor B3 |
787 |
0.64 |
| chr20_3661411_3661562 | 0.26 |
ADAM33 |
ADAM metallopeptidase domain 33 |
1264 |
0.38 |
| chr2_27301899_27302732 | 0.25 |
EMILIN1 |
elastin microfibril interfacer 1 |
880 |
0.32 |
| chr7_100200266_100200615 | 0.25 |
PCOLCE-AS1 |
PCOLCE antisense RNA 1 |
218 |
0.74 |
| chr3_190580567_190581193 | 0.24 |
GMNC |
geminin coiled-coil domain containing |
476 |
0.9 |
| chr19_2720626_2720882 | 0.24 |
DIRAS1 |
DIRAS family, GTP-binding RAS-like 1 |
585 |
0.6 |
| chr13_97899006_97899219 | 0.24 |
MBNL2 |
muscleblind-like splicing regulator 2 |
24503 |
0.25 |
| chr14_103674063_103674327 | 0.24 |
ENSG00000239117 |
. |
4672 |
0.24 |
| chr19_50031610_50031873 | 0.23 |
RCN3 |
reticulocalbin 3, EF-hand calcium binding domain |
142 |
0.87 |
| chr3_44038033_44038921 | 0.23 |
ENSG00000252980 |
. |
74102 |
0.11 |
| chr22_51136634_51136785 | 0.22 |
ENSG00000206841 |
. |
6918 |
0.12 |
| chr2_239754137_239754288 | 0.22 |
TWIST2 |
twist family bHLH transcription factor 2 |
2461 |
0.38 |
| chr8_145925356_145925968 | 0.22 |
AF186192.5 |
|
5257 |
0.15 |
| chr17_59474132_59474398 | 0.22 |
RP11-332H18.4 |
|
1457 |
0.28 |
| chr1_33207592_33208218 | 0.22 |
KIAA1522 |
KIAA1522 |
419 |
0.8 |
| chr13_74315255_74315493 | 0.22 |
KLF12 |
Kruppel-like factor 12 |
253812 |
0.02 |
| chr10_11784750_11785238 | 0.22 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
549 |
0.83 |
| chr4_20255917_20256182 | 0.21 |
SLIT2 |
slit homolog 2 (Drosophila) |
494 |
0.89 |
| chr17_29887977_29888214 | 0.21 |
ENSG00000207614 |
. |
1080 |
0.35 |
| chr4_87517608_87517759 | 0.21 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
837 |
0.62 |
| chr12_77719208_77719557 | 0.21 |
ENSG00000238769 |
. |
162577 |
0.04 |
| chr11_35440407_35440646 | 0.21 |
RP4-683L5.1 |
|
78 |
0.51 |
| chr1_1292304_1292633 | 0.21 |
MXRA8 |
matrix-remodelling associated 8 |
1447 |
0.18 |
| chr7_47576453_47576928 | 0.20 |
TNS3 |
tensin 3 |
2185 |
0.45 |
| chr21_46825103_46825740 | 0.20 |
COL18A1 |
collagen, type XVIII, alpha 1 |
369 |
0.83 |
| chr22_43116992_43117164 | 0.20 |
A4GALT |
alpha 1,4-galactosyltransferase |
25439 |
0.15 |
| chr1_6336062_6336213 | 0.20 |
GPR153 |
G protein-coupled receptor 153 |
15102 |
0.12 |
| chr8_37350646_37351319 | 0.20 |
RP11-150O12.6 |
|
23557 |
0.24 |
| chr16_30996731_30996937 | 0.20 |
HSD3B7 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
297 |
0.67 |
| chr8_48100683_48101124 | 0.19 |
IGLV8OR8-1 |
immunoglobulin lambda variable 8/OR8-1 (pseudogene) |
13617 |
0.23 |
| chr19_49631799_49632178 | 0.19 |
PPFIA3 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3 |
857 |
0.37 |
| chr22_46482810_46483184 | 0.19 |
FLJ27365 |
hsa-mir-4763 |
1114 |
0.31 |
| chr7_143208334_143208676 | 0.19 |
TAS2R41 |
taste receptor, type 2, member 41 |
33539 |
0.13 |
| chr8_6407769_6407977 | 0.19 |
ANGPT2 |
angiopoietin 2 |
12692 |
0.26 |
| chr3_44041432_44041583 | 0.18 |
ENSG00000252980 |
. |
71072 |
0.12 |
| chr19_35522133_35522284 | 0.18 |
SCN1B |
sodium channel, voltage-gated, type I, beta subunit |
483 |
0.65 |
| chr2_110538980_110539399 | 0.18 |
RGPD5 |
RANBP2-like and GRIP domain containing 5 |
11146 |
0.28 |
| chr2_158113918_158114875 | 0.18 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
286 |
0.93 |
| chr19_11305569_11305933 | 0.18 |
KANK2 |
KN motif and ankyrin repeat domains 2 |
117 |
0.94 |
| chr1_14924992_14926197 | 0.17 |
KAZN |
kazrin, periplakin interacting protein |
381 |
0.93 |
| chr14_71605949_71606105 | 0.17 |
PCNX |
pecanex homolog (Drosophila) |
30290 |
0.22 |
| chr15_42749089_42749416 | 0.17 |
ZNF106 |
zinc finger protein 106 |
459 |
0.78 |
| chr10_79397825_79398297 | 0.17 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
66 |
0.98 |
| chr1_19972193_19972357 | 0.17 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
15 |
0.97 |
| chr11_114166754_114166905 | 0.17 |
NNMT |
nicotinamide N-methyltransferase |
282 |
0.93 |
| chr7_5594937_5595088 | 0.17 |
CTB-161C1.1 |
|
1350 |
0.33 |
| chr3_120170199_120170358 | 0.17 |
FSTL1 |
follistatin-like 1 |
178 |
0.97 |
| chr17_15164779_15165041 | 0.17 |
RP11-849N15.1 |
|
578 |
0.55 |
| chr22_43115644_43116713 | 0.17 |
A4GALT |
alpha 1,4-galactosyltransferase |
24539 |
0.15 |
| chr14_38064473_38064843 | 0.17 |
TTC6 |
tetratricopeptide repeat domain 6 |
394 |
0.5 |
| chr11_64427770_64427921 | 0.17 |
AP001092.4 |
|
13974 |
0.14 |
| chr6_6928235_6928478 | 0.17 |
ENSG00000240936 |
. |
10774 |
0.29 |
| chr7_27212262_27212557 | 0.17 |
HOXA10 |
homeobox A10 |
1516 |
0.14 |
| chr15_96888963_96889465 | 0.17 |
ENSG00000222651 |
. |
12724 |
0.15 |
| chr4_187760936_187761144 | 0.17 |
ENSG00000252382 |
. |
17570 |
0.28 |
| chr11_1890233_1890398 | 0.17 |
LSP1 |
lymphocyte-specific protein 1 |
59 |
0.95 |
| chr7_28725956_28726399 | 0.17 |
CREB5 |
cAMP responsive element binding protein 5 |
579 |
0.86 |
| chr19_11466484_11467017 | 0.17 |
DKFZP761J1410 |
Lipid phosphate phosphatase-related protein type 2 |
576 |
0.52 |
| chr19_1154555_1155129 | 0.17 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
293 |
0.85 |
| chr3_104010225_104010492 | 0.17 |
ENSG00000265076 |
. |
236718 |
0.02 |
| chr18_10798656_10798816 | 0.16 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
7088 |
0.29 |
| chr2_200320961_200321297 | 0.16 |
SATB2 |
SATB homeobox 2 |
318 |
0.86 |
| chr19_49370765_49371212 | 0.16 |
PLEKHA4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
136 |
0.91 |
| chr5_80529205_80529404 | 0.16 |
CKMT2 |
creatine kinase, mitochondrial 2 (sarcomeric) |
160 |
0.97 |
| chr9_110349763_110350130 | 0.16 |
ENSG00000202308 |
. |
75581 |
0.1 |
| chr16_3162631_3163041 | 0.16 |
ZNF205 |
zinc finger protein 205 |
40 |
0.8 |
| chr1_7844777_7844986 | 0.16 |
PER3 |
period circadian clock 3 |
118 |
0.95 |
| chr10_119312845_119313011 | 0.16 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
8349 |
0.24 |
| chr16_4313997_4314233 | 0.16 |
TFAP4 |
transcription factor AP-4 (activating enhancer binding protein 4) |
377 |
0.84 |
| chr18_72872404_72872616 | 0.16 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
44958 |
0.18 |
| chr19_6740138_6741070 | 0.16 |
TRIP10 |
thyroid hormone receptor interactor 10 |
325 |
0.81 |
| chr16_863784_863935 | 0.16 |
PRR25 |
proline rich 25 |
8416 |
0.08 |
| chr3_49170609_49171370 | 0.16 |
LAMB2 |
laminin, beta 2 (laminin S) |
438 |
0.69 |
| chr1_215256963_215257162 | 0.16 |
KCNK2 |
potassium channel, subfamily K, member 2 |
202 |
0.97 |
| chr12_41086868_41087222 | 0.16 |
CNTN1 |
contactin 1 |
691 |
0.81 |
| chr2_239768983_239769134 | 0.16 |
TWIST2 |
twist family bHLH transcription factor 2 |
12385 |
0.25 |
| chr3_178336239_178336417 | 0.16 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
59710 |
0.15 |
| chr12_54377889_54378303 | 0.16 |
HOXC-AS3 |
HOXC cluster antisense RNA 3 |
12 |
0.88 |
| chr4_8594931_8595082 | 0.16 |
CPZ |
carboxypeptidase Z |
523 |
0.82 |
| chr1_32045069_32045220 | 0.16 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
3005 |
0.19 |
| chr7_96653221_96653741 | 0.15 |
DLX5 |
distal-less homeobox 5 |
781 |
0.6 |
| chr19_49219826_49219992 | 0.15 |
MAMSTR |
MEF2 activating motif and SAP domain containing transcriptional regulator |
191 |
0.87 |
| chr15_63336117_63336463 | 0.15 |
TPM1 |
tropomyosin 1 (alpha) |
315 |
0.89 |
| chr5_343772_343923 | 0.15 |
AHRR |
aryl-hydrocarbon receptor repressor |
204 |
0.94 |
| chr16_54970178_54971110 | 0.15 |
IRX5 |
iroquois homeobox 5 |
4660 |
0.36 |
| chr18_57127119_57127270 | 0.15 |
RP11-27G24.3 |
|
890 |
0.64 |
| chr19_55684291_55685412 | 0.15 |
SYT5 |
synaptotagmin V |
1083 |
0.28 |
| chr8_93065009_93065160 | 0.15 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
10107 |
0.3 |
| chr21_47407286_47407576 | 0.15 |
COL6A1 |
collagen, type VI, alpha 1 |
5780 |
0.22 |
| chr19_35630347_35630545 | 0.15 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
101 |
0.93 |
| chr19_41224717_41225149 | 0.15 |
ADCK4 |
aarF domain containing kinase 4 |
821 |
0.42 |
| chr1_236227370_236227967 | 0.15 |
NID1 |
nidogen 1 |
741 |
0.67 |
| chr22_51171783_51171934 | 0.15 |
AC000036.4 |
|
4709 |
0.13 |
| chr15_33010201_33010591 | 0.15 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
191 |
0.95 |
| chr5_67830491_67830731 | 0.15 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
242215 |
0.02 |
| chr2_239760169_239760320 | 0.14 |
TWIST2 |
twist family bHLH transcription factor 2 |
3571 |
0.32 |
| chr22_38657243_38657394 | 0.14 |
TMEM184B |
transmembrane protein 184B |
11352 |
0.11 |
| chr20_34025363_34025770 | 0.14 |
GDF5 |
growth differentiation factor 5 |
457 |
0.71 |
| chr2_119587998_119588149 | 0.14 |
EN1 |
engrailed homeobox 1 |
17181 |
0.27 |
| chr6_166419178_166419506 | 0.14 |
ENSG00000252196 |
. |
63513 |
0.12 |
| chr5_170877536_170878064 | 0.14 |
FGF18 |
fibroblast growth factor 18 |
31140 |
0.17 |
| chr3_106917697_106917905 | 0.14 |
ENSG00000220989 |
. |
166228 |
0.04 |
| chr22_22109080_22109472 | 0.14 |
YPEL1 |
yippee-like 1 (Drosophila) |
19153 |
0.12 |
| chr1_84227664_84227815 | 0.14 |
ENSG00000223231 |
. |
31821 |
0.25 |
| chr6_15897253_15897404 | 0.14 |
MYLIP |
myosin regulatory light chain interacting protein |
232028 |
0.02 |
| chr6_23003728_23003960 | 0.14 |
ENSG00000207394 |
. |
121471 |
0.07 |
| chr15_76002333_76002682 | 0.14 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
2682 |
0.14 |
| chr11_70245277_70245996 | 0.14 |
CTTN |
cortactin |
989 |
0.33 |
| chr10_79398316_79398509 | 0.14 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
59 |
0.98 |
| chr6_114181963_114182196 | 0.14 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
3538 |
0.24 |
| chr15_43809873_43810151 | 0.14 |
MAP1A |
microtubule-associated protein 1A |
189 |
0.93 |
| chr15_90371711_90371913 | 0.14 |
ANPEP |
alanyl (membrane) aminopeptidase |
13179 |
0.12 |
| chr7_100765722_100766153 | 0.14 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
4433 |
0.12 |
| chrY_1463557_1463741 | 0.14 |
NA |
NA |
> 106 |
NA |
| chr19_1155345_1155615 | 0.14 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
345 |
0.81 |
| chr17_75474006_75474173 | 0.14 |
SEPT9 |
septin 9 |
2764 |
0.22 |
| chr19_18760778_18760992 | 0.14 |
KLHL26 |
kelch-like family member 26 |
13018 |
0.11 |
| chr2_46557288_46557606 | 0.14 |
EPAS1 |
endothelial PAS domain protein 1 |
32906 |
0.2 |
| chr1_214723618_214723885 | 0.14 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
815 |
0.74 |
| chr4_95649180_95649331 | 0.14 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
29864 |
0.26 |
| chr1_162336853_162337007 | 0.14 |
C1orf226 |
chromosome 1 open reading frame 226 |
232 |
0.86 |
| chr10_128594353_128594780 | 0.14 |
DOCK1 |
dedicator of cytokinesis 1 |
588 |
0.8 |
| chr12_679488_679709 | 0.14 |
RP5-1154L15.2 |
|
16054 |
0.13 |
| chr8_125827951_125828102 | 0.14 |
ENSG00000263735 |
. |
6274 |
0.28 |
| chr17_19066222_19066403 | 0.13 |
AC007952.6 |
Uncharacterized protein |
3888 |
0.1 |
| chr7_127761966_127762314 | 0.13 |
ENSG00000207588 |
. |
40227 |
0.16 |
| chr20_36531075_36531364 | 0.13 |
VSTM2L |
V-set and transmembrane domain containing 2 like |
280 |
0.93 |
| chr16_58534138_58534342 | 0.13 |
NDRG4 |
NDRG family member 4 |
190 |
0.92 |
| chr19_14606640_14606859 | 0.13 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
195 |
0.89 |
| chr19_9473504_9474272 | 0.13 |
ZNF177 |
zinc finger protein 177 |
181 |
0.93 |
| chr19_41073613_41074217 | 0.13 |
SHKBP1 |
SH3KBP1 binding protein 1 |
8842 |
0.14 |
| chr1_212643718_212643913 | 0.13 |
NENF |
neudesin neurotrophic factor |
37586 |
0.14 |
| chr17_881930_882232 | 0.13 |
NXN |
nucleoredoxin |
929 |
0.47 |
| chr6_29974031_29974191 | 0.13 |
HLA-J |
major histocompatibility complex, class I, J (pseudogene) |
272 |
0.88 |
| chr18_65691781_65691932 | 0.13 |
DSEL |
dermatan sulfate epimerase-like |
507639 |
0.0 |
| chr5_126625896_126626411 | 0.13 |
MEGF10 |
multiple EGF-like-domains 10 |
370 |
0.91 |
| chr20_60981442_60981982 | 0.13 |
CABLES2 |
Cdk5 and Abl enzyme substrate 2 |
629 |
0.63 |
| chr2_229046102_229046253 | 0.13 |
SPHKAP |
SPHK1 interactor, AKAP domain containing |
165 |
0.97 |
| chr11_66081550_66081990 | 0.13 |
RP11-867G23.13 |
|
1446 |
0.16 |
| chr17_19118492_19118663 | 0.13 |
EPN2 |
epsin 2 |
351 |
0.77 |
| chr5_77945062_77945285 | 0.13 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
525 |
0.88 |
| chrX_1513557_1513776 | 0.13 |
SLC25A6 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
2049 |
0.26 |
| chr1_227015624_227015805 | 0.13 |
PSEN2 |
presenilin 2 (Alzheimer disease 4) |
42171 |
0.16 |
| chr7_155006231_155006400 | 0.13 |
AC099552.4 |
Uncharacterized protein |
16169 |
0.2 |
| chr12_56115218_56115698 | 0.13 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
1261 |
0.23 |
| chr20_52533519_52533820 | 0.13 |
AC005220.3 |
|
23030 |
0.24 |
| chr8_143782547_143782698 | 0.13 |
LY6K |
lymphocyte antigen 6 complex, locus K |
732 |
0.49 |
| chr5_42423499_42423764 | 0.13 |
GHR |
growth hormone receptor |
248 |
0.96 |
| chr4_189333265_189333416 | 0.13 |
RP11-366H4.3 |
|
268807 |
0.02 |
| chr1_223302857_223303051 | 0.13 |
TLR5 |
toll-like receptor 5 |
5144 |
0.33 |
| chr14_100346663_100346814 | 0.13 |
EML1 |
echinoderm microtubule associated protein like 1 |
14792 |
0.22 |
| chr7_29603979_29604650 | 0.13 |
PRR15 |
proline rich 15 |
752 |
0.51 |
| chr7_72980210_72980413 | 0.13 |
BCL7B |
B-cell CLL/lymphoma 7B |
7979 |
0.14 |
| chr6_108490135_108490714 | 0.13 |
NR2E1 |
nuclear receptor subfamily 2, group E, member 1 |
457 |
0.8 |
| chr19_2579056_2579259 | 0.13 |
CTC-265F19.2 |
|
32703 |
0.12 |
| chr8_57358725_57358876 | 0.13 |
PENK |
proenkephalin |
211 |
0.73 |
| chr11_60928884_60929113 | 0.13 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
91 |
0.97 |
| chr22_28416332_28416483 | 0.13 |
ENSG00000235954 |
. |
22842 |
0.17 |
| chr17_1179389_1179559 | 0.13 |
TUSC5 |
tumor suppressor candidate 5 |
3483 |
0.21 |
| chr7_100816521_100816891 | 0.13 |
NAT16 |
N-acetyltransferase 16 (GCN5-related, putative) |
5811 |
0.1 |
| chr10_100026603_100026754 | 0.13 |
LOXL4 |
lysyl oxidase-like 4 |
1329 |
0.48 |
| chr2_151342879_151343387 | 0.13 |
RND3 |
Rho family GTPase 3 |
1048 |
0.71 |
| chr12_127865548_127865745 | 0.13 |
ENSG00000253089 |
. |
59746 |
0.16 |
| chr8_130472273_130472436 | 0.13 |
ENSG00000263763 |
. |
24034 |
0.12 |
| chr12_52404603_52404754 | 0.12 |
GRASP |
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
289 |
0.86 |
| chr4_188097696_188097847 | 0.12 |
ENSG00000252382 |
. |
319161 |
0.01 |
| chr11_66080835_66080986 | 0.12 |
RP11-867G23.13 |
|
586 |
0.45 |
| chr17_13503597_13503828 | 0.12 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
1532 |
0.51 |
| chr11_9113358_9113598 | 0.12 |
SCUBE2 |
signal peptide, CUB domain, EGF-like 2 |
328 |
0.83 |
| chr9_112259733_112260156 | 0.12 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
642 |
0.76 |
| chrX_11445632_11445915 | 0.12 |
ARHGAP6 |
Rho GTPase activating protein 6 |
120 |
0.98 |
| chr16_86866838_86867009 | 0.12 |
FOXL1 |
forkhead box L1 |
254808 |
0.02 |
| chr11_111784054_111784237 | 0.12 |
CRYAB |
crystallin, alpha B |
74 |
0.89 |
| chr11_65292927_65293752 | 0.12 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
455 |
0.69 |
| chr19_46985200_46985424 | 0.12 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
10492 |
0.13 |
| chr2_236569141_236569292 | 0.12 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
9032 |
0.28 |
| chr12_1905695_1906675 | 0.12 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
14701 |
0.2 |
| chr22_16193199_16193350 | 0.12 |
LL22NC03-N64E9.1 |
|
12270 |
0.18 |
| chr20_30063006_30063157 | 0.12 |
REM1 |
RAS (RAD and GEM)-like GTP-binding 1 |
15 |
0.96 |
| chr8_23201528_23201679 | 0.12 |
ENSG00000253837 |
. |
7882 |
0.17 |
| chr16_69207418_69207583 | 0.12 |
ENSG00000207083 |
. |
2180 |
0.21 |
| chr7_19155713_19156221 | 0.12 |
TWIST1 |
twist family bHLH transcription factor 1 |
1328 |
0.36 |
| chr20_31045078_31045234 | 0.12 |
RP5-1184F4.5 |
|
7598 |
0.18 |
| chr10_49812202_49812558 | 0.12 |
ARHGAP22 |
Rho GTPase activating protein 22 |
617 |
0.77 |
| chr12_57620921_57621410 | 0.12 |
ENSG00000202067 |
. |
592 |
0.53 |
| chr7_96634475_96634654 | 0.12 |
DLX6 |
distal-less homeobox 6 |
296 |
0.79 |
| chr3_131854540_131854927 | 0.12 |
CPNE4 |
copine IV |
96283 |
0.08 |
| chr2_218866425_218866801 | 0.12 |
TNS1 |
tensin 1 |
1105 |
0.51 |
| chr11_67210244_67210509 | 0.12 |
CORO1B |
coronin, actin binding protein, 1B |
567 |
0.48 |
| chr11_64067205_64067580 | 0.12 |
TEX40 |
testis expressed 40 |
471 |
0.54 |
| chr9_35110303_35110495 | 0.12 |
FAM214B |
family with sequence similarity 214, member B |
1021 |
0.37 |
| chr14_33092835_33092986 | 0.12 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
112147 |
0.07 |
| chr9_133482900_133483051 | 0.12 |
FUBP3 |
far upstream element (FUSE) binding protein 3 |
27982 |
0.18 |
| chr6_161664635_161664811 | 0.12 |
AGPAT4 |
1-acylglycerol-3-phosphate O-acyltransferase 4 |
30328 |
0.25 |
| chr17_17110093_17110388 | 0.12 |
PLD6 |
phospholipase D family, member 6 |
611 |
0.61 |
| chr19_11275038_11275287 | 0.12 |
SPC24 |
SPC24, NDC80 kinetochore complex component |
8678 |
0.12 |
| chr5_112823740_112823994 | 0.12 |
MCC |
mutated in colorectal cancers |
660 |
0.69 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.1 | 0.4 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.2 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.3 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.1 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.2 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.1 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.1 | GO:1903579 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0060510 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0014824 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.3 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.2 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.0 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0072283 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.0 | 0.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.0 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.3 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:0042482 | positive regulation of odontogenesis(GO:0042482) positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.0 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.1 | GO:0098876 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.0 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0031946 | regulation of ketone biosynthetic process(GO:0010566) regulation of glucocorticoid biosynthetic process(GO:0031946) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.0 | GO:0015838 | amino-acid betaine transport(GO:0015838) carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0031392 | negative regulation of female receptivity(GO:0007621) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.2 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.0 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.3 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |