Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
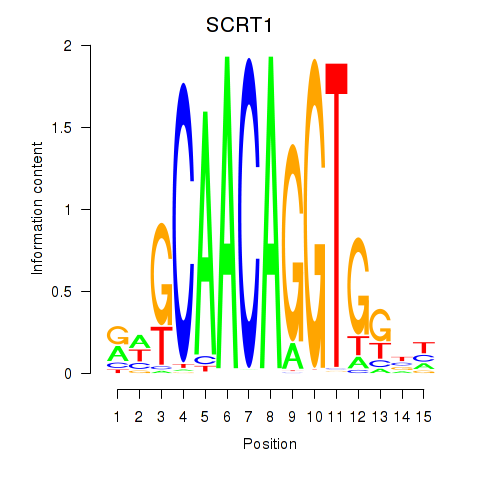
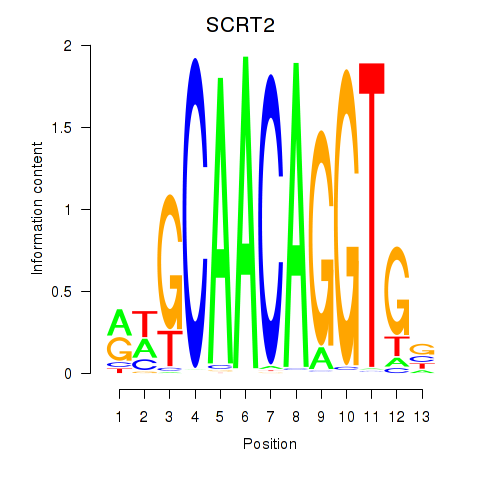
Results for SCRT1_SCRT2
Z-value: 1.20
Transcription factors associated with SCRT1_SCRT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SCRT1
|
ENSG00000170616.9 | scratch family transcriptional repressor 1 |
|
SCRT2
|
ENSG00000215397.3 | scratch family transcriptional repressor 2 |
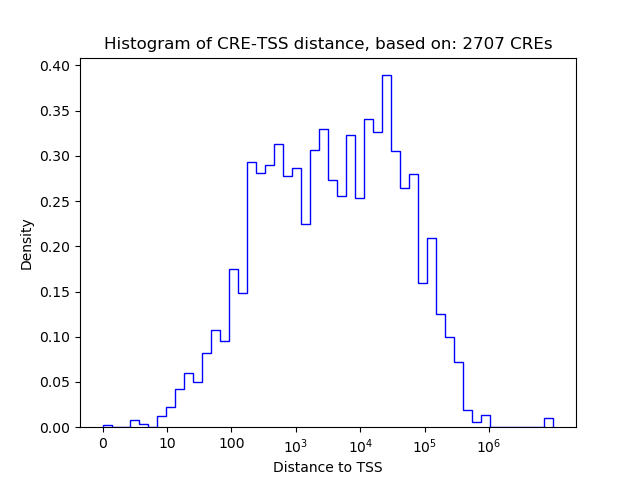
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_145555817_145556064 | SCRT1 | 4003 | 0.086085 | -0.75 | 2.0e-02 | Click! |
| chr8_145560876_145561049 | SCRT1 | 1019 | 0.277140 | 0.52 | 1.5e-01 | Click! |
| chr8_145561219_145561461 | SCRT1 | 1397 | 0.193360 | -0.31 | 4.1e-01 | Click! |
| chr8_145555468_145555619 | SCRT1 | 4400 | 0.082971 | -0.26 | 5.0e-01 | Click! |
| chr8_145562079_145562230 | SCRT1 | 2211 | 0.122634 | -0.14 | 7.3e-01 | Click! |
| chr20_656859_657010 | SCRT2 | 111 | 0.853796 | 0.48 | 1.9e-01 | Click! |
| chr20_657176_657327 | SCRT2 | 428 | 0.659045 | 0.28 | 4.7e-01 | Click! |
Activity of the SCRT1_SCRT2 motif across conditions
Conditions sorted by the z-value of the SCRT1_SCRT2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
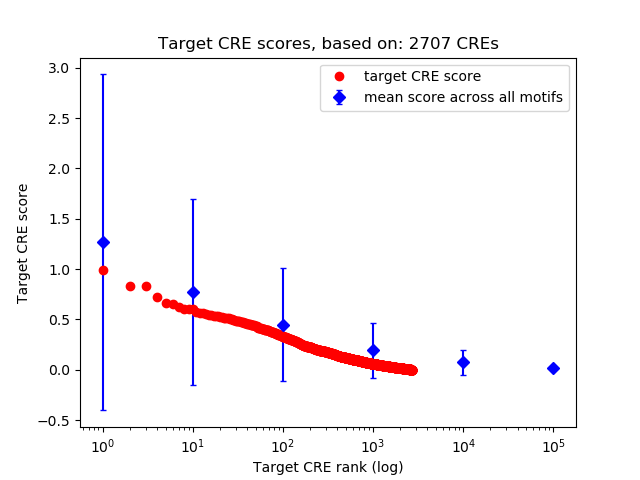
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_112573817_112573968 | 0.99 |
LAMA4 |
laminin, alpha 4 |
1795 |
0.35 |
| chr4_169553568_169553728 | 0.83 |
PALLD |
palladin, cytoskeletal associated protein |
880 |
0.63 |
| chrX_99986862_99987124 | 0.83 |
SYTL4 |
synaptotagmin-like 4 |
114 |
0.97 |
| chr6_121756849_121757212 | 0.72 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
192 |
0.94 |
| chr3_186505500_186505651 | 0.66 |
ENSG00000263776 |
. |
174 |
0.69 |
| chr3_55515212_55515397 | 0.65 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
80 |
0.98 |
| chr5_89854556_89855048 | 0.62 |
GPR98 |
G protein-coupled receptor 98 |
185 |
0.96 |
| chr5_150161983_150162220 | 0.61 |
AC010441.1 |
|
4234 |
0.18 |
| chr20_30300696_30301130 | 0.60 |
AL160175.1 |
|
8088 |
0.13 |
| chr10_63813819_63813970 | 0.60 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
4924 |
0.3 |
| chr3_44038033_44038921 | 0.57 |
ENSG00000252980 |
. |
74102 |
0.11 |
| chr5_138680031_138680182 | 0.56 |
PAIP2 |
poly(A) binding protein interacting protein 2 |
1966 |
0.19 |
| chr20_25213879_25214247 | 0.56 |
AL035252.1 |
HCG2018895; Uncharacterized protein |
6693 |
0.16 |
| chr3_25471508_25471659 | 0.55 |
RARB |
retinoic acid receptor, beta |
1781 |
0.47 |
| chr12_71003154_71004126 | 0.55 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
16 |
0.99 |
| chr20_55206861_55207029 | 0.54 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
1007 |
0.5 |
| chr6_41336919_41337070 | 0.54 |
ENSG00000238867 |
. |
13807 |
0.17 |
| chr9_114671696_114672066 | 0.53 |
UGCG |
UDP-glucose ceramide glucosyltransferase |
12835 |
0.19 |
| chr15_30113385_30113906 | 0.53 |
TJP1 |
tight junction protein 1 |
106 |
0.95 |
| chr7_19144831_19144982 | 0.53 |
AC003986.6 |
|
7191 |
0.17 |
| chr5_15500207_15500358 | 0.52 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
23 |
0.99 |
| chr14_53418107_53419061 | 0.52 |
FERMT2 |
fermitin family member 2 |
569 |
0.83 |
| chr8_122650853_122651057 | 0.51 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
578 |
0.78 |
| chr2_217559691_217559970 | 0.51 |
IGFBP5 |
insulin-like growth factor binding protein 5 |
313 |
0.72 |
| chr3_43995391_43995578 | 0.51 |
ENSG00000252980 |
. |
117095 |
0.06 |
| chr1_215256963_215257162 | 0.51 |
KCNK2 |
potassium channel, subfamily K, member 2 |
202 |
0.97 |
| chr20_36154288_36154588 | 0.51 |
BLCAP |
bladder cancer associated protein |
1485 |
0.45 |
| chr16_2286497_2287265 | 0.49 |
DNASE1L2 |
deoxyribonuclease I-like 2 |
54 |
0.91 |
| chr2_70442433_70442584 | 0.49 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
13942 |
0.14 |
| chrX_128787954_128788123 | 0.49 |
APLN |
apelin |
876 |
0.69 |
| chr3_185677710_185678274 | 0.48 |
RP11-443P15.2 |
|
164 |
0.95 |
| chr5_10306978_10307734 | 0.48 |
CMBL |
carboxymethylenebutenolidase homolog (Pseudomonas) |
782 |
0.53 |
| chr20_43971537_43971915 | 0.48 |
SDC4 |
syndecan 4 |
5338 |
0.13 |
| chr9_4339562_4339725 | 0.48 |
ENSG00000221730 |
. |
38851 |
0.13 |
| chr4_41157747_41157945 | 0.48 |
ENSG00000207198 |
. |
41887 |
0.14 |
| chr2_101437792_101437972 | 0.48 |
NPAS2 |
neuronal PAS domain protein 2 |
395 |
0.8 |
| chr6_42109948_42110375 | 0.47 |
C6orf132 |
chromosome 6 open reading frame 132 |
21 |
0.97 |
| chr11_27743435_27743805 | 0.46 |
BDNF |
brain-derived neurotrophic factor |
15 |
0.99 |
| chr8_58660509_58660660 | 0.46 |
ENSG00000252057 |
. |
191440 |
0.03 |
| chr6_130689068_130689442 | 0.46 |
TMEM200A |
transmembrane protein 200A |
2376 |
0.3 |
| chr6_164616629_164616780 | 0.45 |
ENSG00000266128 |
. |
355111 |
0.01 |
| chr4_177365034_177365248 | 0.45 |
RP11-87F15.2 |
|
123532 |
0.05 |
| chr5_32712676_32712984 | 0.45 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
393 |
0.9 |
| chr8_104155072_104155223 | 0.45 |
C8orf56 |
chromosome 8 open reading frame 56 |
1444 |
0.27 |
| chr12_3232327_3232575 | 0.45 |
TSPAN9-IT1 |
TSPAN9 intronic transcript 1 (non-protein coding) |
26512 |
0.19 |
| chr19_46974752_46975184 | 0.45 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
148 |
0.94 |
| chr10_27547544_27548544 | 0.44 |
ARMC4P1 |
armadillo repeat containing 4 pseudogene 1 |
3842 |
0.17 |
| chr22_45900582_45901194 | 0.44 |
FBLN1 |
fibulin 1 |
1962 |
0.36 |
| chr18_61616540_61617060 | 0.44 |
HMSD |
histocompatibility (minor) serpin domain containing |
265 |
0.9 |
| chr12_16759185_16759513 | 0.43 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
116 |
0.98 |
| chr1_95392942_95393358 | 0.43 |
RP4-639F20.1 |
|
28 |
0.82 |
| chr4_148402515_148403207 | 0.43 |
EDNRA |
endothelin receptor type A |
777 |
0.76 |
| chr1_60599792_60599943 | 0.42 |
C1orf87 |
chromosome 1 open reading frame 87 |
60425 |
0.15 |
| chr5_111090218_111090459 | 0.42 |
NREP |
neuronal regeneration related protein |
1610 |
0.42 |
| chr4_101111713_101112234 | 0.41 |
DDIT4L |
DNA-damage-inducible transcript 4-like |
34 |
0.95 |
| chr10_23216488_23217262 | 0.41 |
ARMC3 |
armadillo repeat containing 3 |
78 |
0.98 |
| chr5_137474933_137475144 | 0.41 |
NME5 |
NME/NM23 family member 5 |
66 |
0.95 |
| chr8_77316107_77316384 | 0.41 |
ENSG00000222231 |
. |
138278 |
0.05 |
| chr18_52495652_52496658 | 0.41 |
RAB27B |
RAB27B, member RAS oncogene family |
725 |
0.77 |
| chr11_73022764_73022915 | 0.41 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
229 |
0.89 |
| chr11_93885194_93885476 | 0.41 |
PANX1 |
pannexin 1 |
23240 |
0.23 |
| chr4_41258452_41258603 | 0.40 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
97 |
0.66 |
| chr13_42490447_42490670 | 0.40 |
ENSG00000206962 |
. |
11304 |
0.24 |
| chr1_220101375_220102142 | 0.40 |
SLC30A10 |
solute carrier family 30, member 10 |
186 |
0.95 |
| chr5_59893443_59893927 | 0.40 |
DEPDC1B |
DEP domain containing 1B |
102276 |
0.07 |
| chr6_11196915_11197406 | 0.39 |
RP3-510L9.1 |
|
23475 |
0.18 |
| chr1_151962822_151962989 | 0.39 |
S100A10 |
S100 calcium binding protein A10 |
2149 |
0.24 |
| chr12_120935516_120935667 | 0.39 |
DYNLL1 |
dynein, light chain, LC8-type 1 |
1675 |
0.19 |
| chr5_155108981_155109215 | 0.39 |
ENSG00000200275 |
. |
163349 |
0.04 |
| chr6_129820127_129820286 | 0.39 |
RP1-69D17.3 |
|
17651 |
0.26 |
| chr3_179754024_179754473 | 0.38 |
PEX5L |
peroxisomal biogenesis factor 5-like |
310 |
0.94 |
| chr2_145408788_145408939 | 0.38 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
130242 |
0.06 |
| chr16_54431828_54431986 | 0.38 |
IRX3 |
iroquois homeobox 3 |
111232 |
0.07 |
| chr1_244394727_244395045 | 0.38 |
C1orf100 |
chromosome 1 open reading frame 100 |
121051 |
0.05 |
| chr17_13502306_13502495 | 0.37 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
2844 |
0.36 |
| chr3_185482197_185482362 | 0.37 |
ENSG00000265470 |
. |
3413 |
0.28 |
| chr2_135812118_135812269 | 0.37 |
RAB3GAP1 |
RAB3 GTPase activating protein subunit 1 (catalytic) |
2324 |
0.27 |
| chr2_192111060_192111278 | 0.37 |
MYO1B |
myosin IB |
160 |
0.97 |
| chr9_107968732_107968883 | 0.36 |
SLC44A1 |
solute carrier family 44 (choline transporter), member 1 |
38096 |
0.21 |
| chr8_141248134_141248335 | 0.36 |
TRAPPC9 |
trafficking protein particle complex 9 |
212795 |
0.02 |
| chr10_9751707_9751888 | 0.36 |
ENSG00000200849 |
. |
522777 |
0.0 |
| chr4_184021000_184021241 | 0.36 |
WWC2 |
WW and C2 domain containing 2 |
475 |
0.85 |
| chr9_94245349_94245513 | 0.36 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
59287 |
0.15 |
| chrX_139584851_139585024 | 0.36 |
SOX3 |
SRY (sex determining region Y)-box 3 |
2288 |
0.43 |
| chr6_27440042_27440937 | 0.36 |
ZNF184 |
zinc finger protein 184 |
29 |
0.98 |
| chr3_19232179_19232330 | 0.36 |
KCNH8 |
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
42308 |
0.21 |
| chrX_45612401_45612855 | 0.36 |
ENSG00000207725 |
. |
6098 |
0.25 |
| chr4_73184982_73185292 | 0.35 |
RP11-373J21.1 |
|
3608 |
0.39 |
| chr10_77190951_77191328 | 0.35 |
RP11-399K21.10 |
|
207 |
0.95 |
| chr1_1552204_1552355 | 0.35 |
MIB2 |
mindbomb E3 ubiquitin protein ligase 2 |
509 |
0.54 |
| chr8_49320680_49320831 | 0.35 |
ENSG00000252710 |
. |
100165 |
0.08 |
| chr3_107280996_107281147 | 0.34 |
BBX |
bobby sox homolog (Drosophila) |
36822 |
0.23 |
| chr9_110248552_110248703 | 0.34 |
KLF4 |
Kruppel-like factor 4 (gut) |
2784 |
0.3 |
| chr16_80716241_80716392 | 0.34 |
ENSG00000265341 |
. |
16856 |
0.18 |
| chr4_187644173_187644789 | 0.34 |
FAT1 |
FAT atypical cadherin 1 |
528 |
0.87 |
| chr20_36780874_36781212 | 0.34 |
TGM2 |
transglutaminase 2 |
12629 |
0.19 |
| chr18_59993646_59993797 | 0.34 |
TNFRSF11A |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
1173 |
0.44 |
| chr3_99356455_99356606 | 0.33 |
COL8A1 |
collagen, type VIII, alpha 1 |
789 |
0.74 |
| chr20_57425359_57425924 | 0.33 |
GNAS-AS1 |
GNAS antisense RNA 1 |
292 |
0.85 |
| chr6_128897010_128897161 | 0.33 |
ENSG00000206982 |
. |
8450 |
0.2 |
| chr5_106724658_106724809 | 0.33 |
EFNA5 |
ephrin-A5 |
281595 |
0.01 |
| chr16_66638822_66640143 | 0.33 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
58 |
0.95 |
| chr1_172106062_172106431 | 0.33 |
ENSG00000207949 |
. |
1801 |
0.34 |
| chr9_21683236_21683387 | 0.33 |
ENSG00000244230 |
. |
16002 |
0.23 |
| chr4_187491923_187492195 | 0.32 |
MTNR1A |
melatonin receptor 1A |
15338 |
0.17 |
| chr9_12776208_12776436 | 0.32 |
LURAP1L |
leucine rich adaptor protein 1-like |
1302 |
0.47 |
| chr10_18629419_18629841 | 0.32 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
36 |
0.99 |
| chr5_127872787_127873476 | 0.32 |
FBN2 |
fibrillin 2 |
383 |
0.63 |
| chr9_100390326_100390477 | 0.32 |
TSTD2 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 |
5456 |
0.17 |
| chr19_35605607_35605885 | 0.32 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
986 |
0.35 |
| chr18_10791509_10791660 | 0.32 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
64 |
0.98 |
| chr3_123418520_123418671 | 0.31 |
MYLK |
myosin light chain kinase |
7369 |
0.19 |
| chr11_122049732_122050124 | 0.31 |
ENSG00000207994 |
. |
26912 |
0.16 |
| chr3_62860670_62860829 | 0.31 |
CADPS |
Ca++-dependent secretion activator |
45 |
0.99 |
| chr8_26152500_26152651 | 0.31 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
1576 |
0.5 |
| chr17_10101159_10101987 | 0.31 |
GAS7 |
growth arrest-specific 7 |
295 |
0.92 |
| chr3_100711576_100712203 | 0.31 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
408 |
0.89 |
| chr4_124476505_124476726 | 0.31 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
155492 |
0.04 |
| chr17_60173604_60173902 | 0.31 |
ENSG00000207123 |
. |
25928 |
0.16 |
| chr10_35645565_35645754 | 0.31 |
CCNY |
cyclin Y |
19857 |
0.19 |
| chr2_23840040_23840191 | 0.30 |
KLHL29 |
kelch-like family member 29 |
25147 |
0.25 |
| chr14_70059777_70059928 | 0.30 |
KIAA0247 |
KIAA0247 |
18461 |
0.2 |
| chr2_70528224_70529180 | 0.30 |
AC022201.5 |
|
119 |
0.81 |
| chr2_173330084_173330561 | 0.30 |
AC078883.3 |
|
418 |
0.83 |
| chr5_72411007_72411158 | 0.30 |
TMEM171 |
transmembrane protein 171 |
5037 |
0.19 |
| chr1_155616210_155616471 | 0.30 |
YY1AP1 |
YY1 associated protein 1 |
30122 |
0.11 |
| chr10_3849010_3849161 | 0.30 |
KLF6 |
Kruppel-like factor 6 |
21612 |
0.22 |
| chr16_67896952_67897145 | 0.29 |
EDC4 |
enhancer of mRNA decapping 4 |
9878 |
0.08 |
| chr18_74843262_74844680 | 0.29 |
MBP |
myelin basic protein |
331 |
0.94 |
| chr2_165811625_165811865 | 0.29 |
SLC38A11 |
solute carrier family 38, member 11 |
11 |
0.98 |
| chr7_119913514_119913995 | 0.29 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
32 |
0.99 |
| chr4_20256931_20257082 | 0.29 |
SLIT2 |
slit homolog 2 (Drosophila) |
463 |
0.9 |
| chr18_76746162_76746313 | 0.29 |
SALL3 |
spalt-like transcription factor 3 |
5445 |
0.31 |
| chr13_95619763_95620412 | 0.29 |
ENSG00000252335 |
. |
51402 |
0.18 |
| chr17_68165729_68165994 | 0.29 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
185 |
0.66 |
| chr1_38059421_38059572 | 0.28 |
GNL2 |
guanine nucleotide binding protein-like 2 (nucleolar) |
2026 |
0.26 |
| chr14_69727713_69728275 | 0.28 |
GALNT16 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
986 |
0.63 |
| chrX_5651570_5651721 | 0.28 |
ENSG00000201660 |
. |
323482 |
0.01 |
| chr17_40162554_40162705 | 0.28 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
664 |
0.41 |
| chr16_4349380_4349548 | 0.28 |
GLIS2 |
GLIS family zinc finger 2 |
15298 |
0.13 |
| chr11_64002356_64003048 | 0.27 |
VEGFB |
vascular endothelial growth factor B |
410 |
0.51 |
| chr11_63581191_63582006 | 0.27 |
C11orf84 |
chromosome 11 open reading frame 84 |
738 |
0.6 |
| chr17_80354838_80354989 | 0.27 |
RP13-20L14.4 |
|
4345 |
0.1 |
| chr21_45123118_45123419 | 0.27 |
PDXK |
pyridoxal (pyridoxine, vitamin B6) kinase |
15707 |
0.17 |
| chr3_10487191_10487342 | 0.27 |
ATP2B2 |
ATPase, Ca++ transporting, plasma membrane 2 |
4031 |
0.22 |
| chr5_128796123_128796371 | 0.27 |
ADAMTS19-AS1 |
ADAMTS19 antisense RNA 1 |
135 |
0.71 |
| chr18_56888364_56888553 | 0.27 |
GRP |
gastrin-releasing peptide |
960 |
0.63 |
| chr11_7273101_7273359 | 0.27 |
SYT9 |
synaptotagmin IX |
49 |
0.98 |
| chr16_9204891_9205042 | 0.27 |
RP11-473I1.10 |
|
3868 |
0.18 |
| chr14_105886805_105887920 | 0.26 |
MTA1 |
metastasis associated 1 |
1087 |
0.31 |
| chr10_24497729_24498381 | 0.26 |
KIAA1217 |
KIAA1217 |
38 |
0.99 |
| chr19_19574756_19575086 | 0.26 |
GATAD2A |
GATA zinc finger domain containing 2A |
570 |
0.67 |
| chr11_61597292_61597443 | 0.26 |
FADS1 |
fatty acid desaturase 1 |
577 |
0.57 |
| chr12_21652771_21652922 | 0.26 |
RECQL |
RecQ protein-like (DNA helicase Q1-like) |
1639 |
0.29 |
| chr6_1873132_1873283 | 0.26 |
FOXC1 |
forkhead box C1 |
262526 |
0.02 |
| chr15_62737317_62737509 | 0.26 |
TLN2 |
talin 2 |
116151 |
0.06 |
| chr15_57508769_57508920 | 0.25 |
TCF12 |
transcription factor 12 |
2784 |
0.37 |
| chr4_126311427_126311944 | 0.25 |
FAT4 |
FAT atypical cadherin 4 |
3406 |
0.33 |
| chr17_66596046_66596347 | 0.25 |
FAM20A |
family with sequence similarity 20, member A |
1334 |
0.52 |
| chr5_151098113_151098265 | 0.25 |
ENSG00000222102 |
. |
14050 |
0.14 |
| chr12_2340763_2340914 | 0.25 |
CACNA1C-AS4 |
CACNA1C antisense RNA 4 |
8193 |
0.25 |
| chr3_27707961_27708112 | 0.25 |
EOMES |
eomesodermin |
55767 |
0.15 |
| chr1_67029534_67029780 | 0.25 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
29692 |
0.2 |
| chr14_92968443_92968594 | 0.25 |
RIN3 |
Ras and Rab interactor 3 |
11600 |
0.27 |
| chr21_46825742_46826418 | 0.25 |
COL18A1 |
collagen, type XVIII, alpha 1 |
1028 |
0.47 |
| chr6_5941079_5941352 | 0.24 |
ENSG00000239472 |
. |
22138 |
0.24 |
| chr8_114446989_114447172 | 0.24 |
CSMD3 |
CUB and Sushi multiple domains 3 |
2032 |
0.42 |
| chr18_47087818_47088752 | 0.24 |
LIPG |
lipase, endothelial |
116 |
0.96 |
| chr20_2114530_2114681 | 0.24 |
ENSG00000263452 |
. |
23322 |
0.19 |
| chr16_12896672_12896922 | 0.24 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
911 |
0.59 |
| chr19_408624_409082 | 0.24 |
C2CD4C |
C2 calcium-dependent domain containing 4C |
286 |
0.86 |
| chr14_23775554_23775975 | 0.24 |
BCL2L2 |
BCL2-like 2 |
207 |
0.54 |
| chr10_17104740_17104895 | 0.24 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
65019 |
0.12 |
| chr17_62675596_62675747 | 0.24 |
SMURF2 |
SMAD specific E3 ubiquitin protein ligase 2 |
17485 |
0.2 |
| chr4_157893196_157893594 | 0.24 |
PDGFC |
platelet derived growth factor C |
849 |
0.66 |
| chr9_94586642_94586793 | 0.24 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
124445 |
0.06 |
| chr7_93943307_93943458 | 0.24 |
COL1A2 |
collagen, type I, alpha 2 |
80491 |
0.11 |
| chr5_58858854_58859005 | 0.24 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
23290 |
0.28 |
| chr1_24645756_24645907 | 0.23 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
19 |
0.97 |
| chr4_74316335_74316542 | 0.23 |
AFP |
alpha-fetoprotein |
14474 |
0.2 |
| chr3_175388674_175388825 | 0.23 |
ENSG00000201648 |
. |
61482 |
0.12 |
| chr13_96742947_96743243 | 0.23 |
HS6ST3 |
heparan sulfate 6-O-sulfotransferase 3 |
2 |
0.99 |
| chr18_67955480_67955943 | 0.23 |
SOCS6 |
suppressor of cytokine signaling 6 |
426 |
0.88 |
| chr1_243518007_243518236 | 0.23 |
ENSG00000265201 |
. |
8643 |
0.26 |
| chr17_55945252_55945551 | 0.23 |
CUEDC1 |
CUE domain containing 1 |
625 |
0.71 |
| chr1_15527398_15527585 | 0.23 |
C1orf195 |
chromosome 1 open reading frame 195 |
29678 |
0.16 |
| chr5_78053562_78053713 | 0.23 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
108989 |
0.07 |
| chr9_16425913_16426064 | 0.23 |
BNC2 |
basonuclin 2 |
10382 |
0.27 |
| chr8_62646329_62646480 | 0.23 |
ENSG00000264408 |
. |
19057 |
0.22 |
| chr17_74696523_74696739 | 0.23 |
ENSG00000212418 |
. |
3178 |
0.11 |
| chr12_78429661_78429970 | 0.23 |
RP11-136F16.1 |
|
55702 |
0.16 |
| chr12_75784171_75784441 | 0.23 |
CAPS2 |
calcyphosine 2 |
375 |
0.6 |
| chr5_174115741_174115892 | 0.23 |
MSX2 |
msh homeobox 2 |
35720 |
0.21 |
| chr18_56888101_56888263 | 0.23 |
GRP |
gastrin-releasing peptide |
684 |
0.75 |
| chr4_141348226_141349120 | 0.23 |
CLGN |
calmegin |
91 |
0.97 |
| chrX_130897762_130897913 | 0.23 |
ENSG00000200587 |
. |
175147 |
0.03 |
| chr16_50401597_50402736 | 0.23 |
RP11-21B23.1 |
|
325 |
0.64 |
| chr10_33630285_33630504 | 0.23 |
NRP1 |
neuropilin 1 |
5204 |
0.3 |
| chr5_39102785_39103048 | 0.23 |
AC008964.1 |
|
2442 |
0.36 |
| chr12_71113512_71113735 | 0.23 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
34750 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.2 | 0.7 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.6 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.6 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.1 | 0.3 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.3 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.3 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.3 | GO:0014820 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.1 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.1 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.4 | GO:0006183 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.1 | 0.4 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.9 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0060433 | bronchus development(GO:0060433) |
| 0.0 | 0.0 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.2 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.2 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.3 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.5 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.2 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.2 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.6 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0060008 | Sertoli cell differentiation(GO:0060008) |
| 0.0 | 0.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.3 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.5 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.0 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.4 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0001542 | ovulation from ovarian follicle(GO:0001542) |
| 0.0 | 0.3 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.5 | GO:0006921 | cellular component disassembly involved in execution phase of apoptosis(GO:0006921) execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.1 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.0 | 0.0 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.0 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.1 | 0.4 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.4 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.8 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.4 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.4 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.0 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 1.1 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.0 | GO:0045277 | mitochondrial respiratory chain complex IV(GO:0005751) respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.6 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.1 | 0.3 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.7 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.4 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.1 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.1 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |