Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
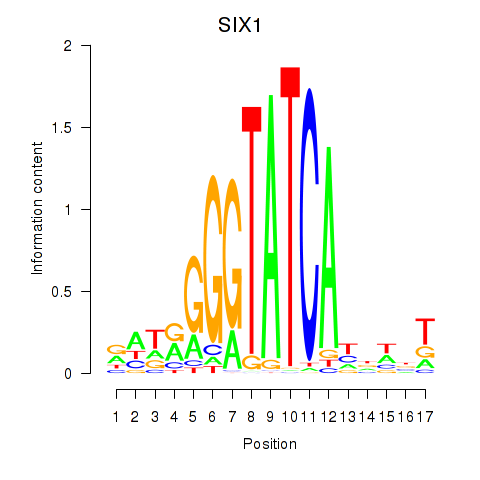
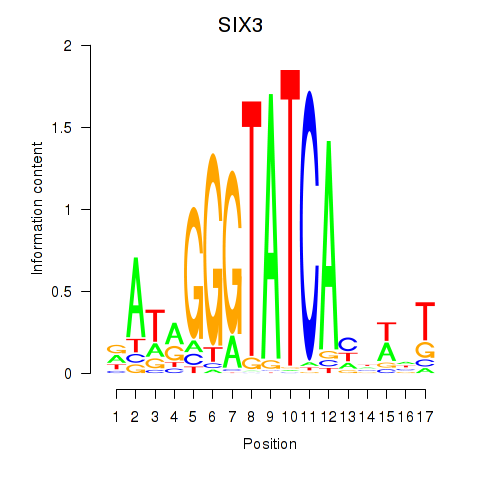
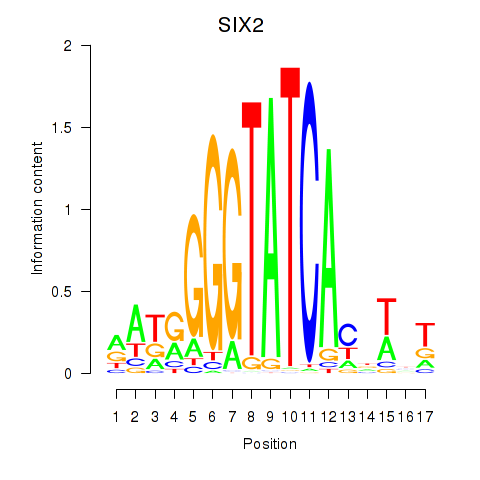
Results for SIX1_SIX3_SIX2
Z-value: 1.68
Transcription factors associated with SIX1_SIX3_SIX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX1
|
ENSG00000126778.7 | SIX homeobox 1 |
|
SIX3
|
ENSG00000138083.3 | SIX homeobox 3 |
|
SIX2
|
ENSG00000170577.7 | SIX homeobox 2 |
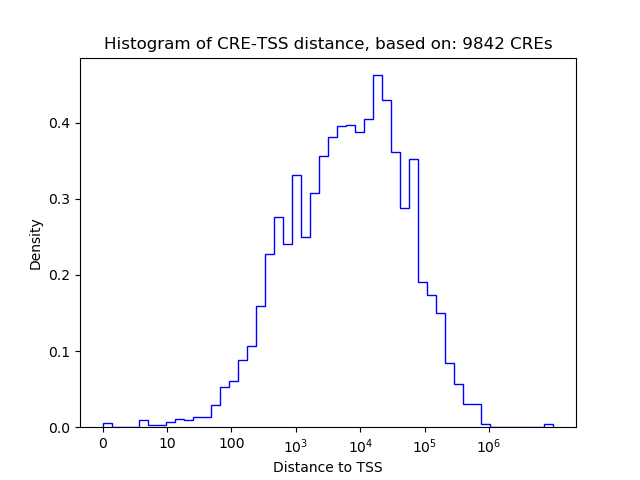
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_61109318_61109469 | SIX1 | 6787 | 0.224023 | -0.82 | 6.5e-03 | Click! |
| chr14_61113971_61114451 | SIX1 | 1969 | 0.367365 | -0.73 | 2.5e-02 | Click! |
| chr14_61116322_61116473 | SIX1 | 217 | 0.946641 | -0.72 | 3.0e-02 | Click! |
| chr14_61123462_61124719 | SIX1 | 887 | 0.641189 | -0.68 | 4.4e-02 | Click! |
| chr14_61116520_61116671 | SIX1 | 415 | 0.866959 | -0.67 | 4.9e-02 | Click! |
| chr2_45241005_45241270 | SIX2 | 4568 | 0.304529 | -0.71 | 3.2e-02 | Click! |
| chr2_45235607_45236412 | SIX2 | 560 | 0.835356 | -0.70 | 3.6e-02 | Click! |
| chr2_45236575_45236726 | SIX2 | 81 | 0.982179 | -0.69 | 3.9e-02 | Click! |
| chr2_45405843_45405994 | SIX2 | 169349 | 0.035597 | -0.69 | 4.0e-02 | Click! |
| chr2_45395914_45396553 | SIX2 | 159664 | 0.039346 | -0.67 | 4.9e-02 | Click! |
| chr2_45168726_45169073 | SIX3 | 3 | 0.605121 | 0.07 | 8.6e-01 | Click! |
Activity of the SIX1_SIX3_SIX2 motif across conditions
Conditions sorted by the z-value of the SIX1_SIX3_SIX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
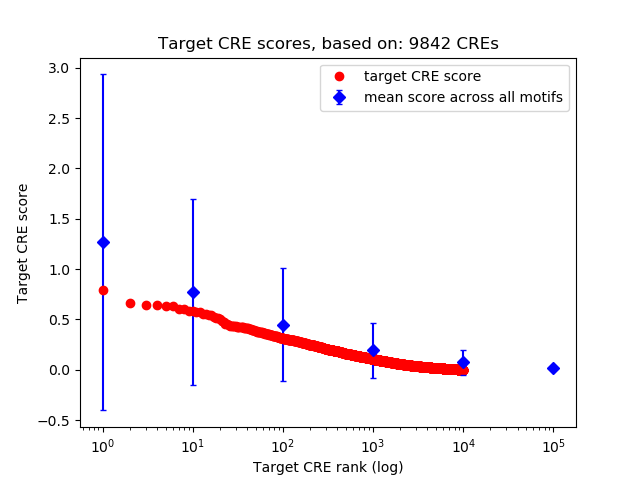
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_42056193_42056486 | 0.80 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
453 |
0.8 |
| chr2_106775791_106775996 | 0.66 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
1147 |
0.57 |
| chr12_29255881_29256032 | 0.64 |
ENSG00000222481 |
. |
17859 |
0.25 |
| chr14_22949288_22949494 | 0.64 |
ENSG00000251002 |
. |
887 |
0.35 |
| chr6_26442034_26442281 | 0.64 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
1371 |
0.29 |
| chr4_77910622_77910773 | 0.63 |
SEPT11 |
septin 11 |
4520 |
0.28 |
| chr6_76203816_76203967 | 0.61 |
FILIP1 |
filamin A interacting protein 1 |
437 |
0.83 |
| chr6_151562009_151562901 | 0.60 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
946 |
0.51 |
| chr2_208390303_208390479 | 0.59 |
CREB1 |
cAMP responsive element binding protein 1 |
4070 |
0.24 |
| chr20_62643605_62643756 | 0.59 |
ZNF512B |
zinc finger protein 512B |
26258 |
0.07 |
| chrX_13268074_13268225 | 0.58 |
GS1-600G8.3 |
|
60622 |
0.13 |
| chr20_62643970_62644121 | 0.57 |
ZNF512B |
zinc finger protein 512B |
25893 |
0.07 |
| chr7_5728695_5728919 | 0.56 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
8715 |
0.21 |
| chr17_74749298_74749501 | 0.56 |
ENSG00000200257 |
. |
405 |
0.71 |
| chr8_29958413_29958564 | 0.54 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
968 |
0.49 |
| chr16_84802144_84802295 | 0.54 |
USP10 |
ubiquitin specific peptidase 10 |
346 |
0.89 |
| chr3_50645256_50645407 | 0.53 |
CISH |
cytokine inducible SH2-containing protein |
3872 |
0.17 |
| chr6_128255928_128256079 | 0.52 |
THEMIS |
thymocyte selection associated |
16227 |
0.26 |
| chr1_210004793_210004944 | 0.51 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
3516 |
0.21 |
| chrX_101409617_101409776 | 0.51 |
BEX5 |
brain expressed, X-linked 5 |
1066 |
0.45 |
| chr7_130637471_130637622 | 0.48 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
31322 |
0.2 |
| chr2_158296564_158297076 | 0.48 |
CYTIP |
cytohesin 1 interacting protein |
894 |
0.5 |
| chr12_47604790_47604941 | 0.46 |
PCED1B |
PC-esterase domain containing 1B |
5187 |
0.24 |
| chr2_219761502_219762270 | 0.45 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
15003 |
0.11 |
| chr4_56276034_56276185 | 0.44 |
ENSG00000239040 |
. |
2694 |
0.22 |
| chr14_23013027_23013178 | 0.44 |
AE000662.92 |
Uncharacterized protein |
12432 |
0.1 |
| chr4_40178201_40178376 | 0.44 |
RP11-395I6.3 |
|
9993 |
0.19 |
| chr7_130627058_130627209 | 0.44 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
41735 |
0.17 |
| chr14_22695818_22695969 | 0.43 |
ENSG00000238634 |
. |
85006 |
0.09 |
| chrX_15756367_15757484 | 0.43 |
CA5B |
carbonic anhydrase VB, mitochondrial |
532 |
0.77 |
| chr4_113912419_113912570 | 0.43 |
RP11-650J17.2 |
|
43213 |
0.15 |
| chr10_125828390_125828541 | 0.43 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
22224 |
0.25 |
| chr7_150216886_150217185 | 0.43 |
GIMAP7 |
GTPase, IMAP family member 7 |
5117 |
0.21 |
| chr5_40686354_40686614 | 0.43 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
6884 |
0.22 |
| chr9_77707832_77707983 | 0.43 |
OSTF1 |
osteoclast stimulating factor 1 |
4448 |
0.21 |
| chr11_29141848_29142035 | 0.42 |
ENSG00000211499 |
. |
156295 |
0.04 |
| chr6_125064729_125064880 | 0.42 |
RNF217 |
ring finger protein 217 |
218887 |
0.02 |
| chr12_24967868_24968019 | 0.42 |
RP11-625L16.3 |
|
15412 |
0.24 |
| chr1_198614639_198614833 | 0.41 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
6444 |
0.26 |
| chr9_134145569_134145886 | 0.41 |
FAM78A |
family with sequence similarity 78, member A |
153 |
0.95 |
| chr10_116063923_116064706 | 0.41 |
VWA2 |
von Willebrand factor A domain containing 2 |
65225 |
0.11 |
| chr5_139502851_139503002 | 0.41 |
IGIP |
IgA-inducing protein |
2595 |
0.21 |
| chr15_52548772_52549093 | 0.40 |
ENSG00000221052 |
. |
20465 |
0.17 |
| chr9_115811998_115812149 | 0.40 |
ZFP37 |
ZFP37 zinc finger protein |
6895 |
0.27 |
| chr9_92786212_92786363 | 0.40 |
ENSG00000263967 |
. |
470 |
0.9 |
| chr12_57527559_57527864 | 0.40 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
1789 |
0.2 |
| chr17_75373020_75373171 | 0.40 |
RP11-936I5.1 |
|
223 |
0.86 |
| chr11_128911246_128911397 | 0.39 |
ENSG00000206847 |
. |
10967 |
0.19 |
| chr19_39728332_39728587 | 0.39 |
IFNL3 |
interferon, lambda 3 |
7187 |
0.12 |
| chr6_143264256_143264520 | 0.39 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
1950 |
0.43 |
| chr1_111210725_111210963 | 0.38 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
6811 |
0.19 |
| chr6_136845943_136846225 | 0.38 |
MAP7 |
microtubule-associated protein 7 |
1037 |
0.53 |
| chr1_84630171_84630563 | 0.37 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
3 |
0.99 |
| chr4_143768164_143768496 | 0.37 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
255 |
0.96 |
| chr1_111211265_111211483 | 0.37 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
6281 |
0.19 |
| chr3_56949638_56949992 | 0.37 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
684 |
0.75 |
| chr5_107438203_107438354 | 0.37 |
FBXL17 |
F-box and leucine-rich repeat protein 17 |
265377 |
0.02 |
| chr1_186291918_186292069 | 0.37 |
ENSG00000202025 |
. |
10933 |
0.17 |
| chr20_35258352_35258894 | 0.37 |
SLA2 |
Src-like-adaptor 2 |
15661 |
0.12 |
| chr3_152077898_152078049 | 0.37 |
TMEM14E |
transmembrane protein 14E |
19194 |
0.22 |
| chr9_131318552_131318703 | 0.37 |
SPTAN1 |
spectrin, alpha, non-erythrocytic 1 |
3758 |
0.14 |
| chr19_41813326_41813520 | 0.37 |
CCDC97 |
coiled-coil domain containing 97 |
2671 |
0.16 |
| chr10_60336470_60336621 | 0.37 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
63645 |
0.15 |
| chr2_175710164_175710315 | 0.36 |
CHN1 |
chimerin 1 |
894 |
0.68 |
| chr2_182321276_182321427 | 0.36 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
583 |
0.85 |
| chr4_129732983_129734245 | 0.36 |
JADE1 |
jade family PHD finger 1 |
615 |
0.83 |
| chr9_95474388_95474658 | 0.36 |
IPPK |
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
41976 |
0.14 |
| chr16_68773930_68774081 | 0.36 |
ENSG00000200558 |
. |
2378 |
0.23 |
| chr14_22783453_22783604 | 0.35 |
ENSG00000251002 |
. |
118191 |
0.04 |
| chr6_45463009_45463160 | 0.35 |
RUNX2 |
runt-related transcription factor 2 |
72862 |
0.11 |
| chr12_6744335_6744486 | 0.35 |
LPAR5 |
lysophosphatidic acid receptor 5 |
1203 |
0.25 |
| chr2_143887292_143887566 | 0.35 |
ARHGAP15 |
Rho GTPase activating protein 15 |
546 |
0.84 |
| chr10_28621171_28621410 | 0.35 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
2125 |
0.36 |
| chr1_61332097_61332364 | 0.35 |
NFIA |
nuclear factor I/A |
1299 |
0.6 |
| chr14_61802494_61802645 | 0.34 |
PRKCH |
protein kinase C, eta |
8391 |
0.2 |
| chr20_43110715_43110866 | 0.34 |
TTPAL |
tocopherol (alpha) transfer protein-like |
6230 |
0.13 |
| chr9_110745973_110746124 | 0.34 |
ENSG00000222459 |
. |
64789 |
0.14 |
| chr6_33378848_33379665 | 0.34 |
PHF1 |
PHD finger protein 1 |
518 |
0.64 |
| chr2_106355418_106355569 | 0.34 |
NCK2 |
NCK adaptor protein 2 |
5861 |
0.32 |
| chr10_126430313_126430464 | 0.34 |
FAM53B |
family with sequence similarity 53, member B |
1151 |
0.46 |
| chr4_119748782_119749072 | 0.34 |
SEC24D |
SEC24 family member D |
8366 |
0.28 |
| chr9_130461454_130462084 | 0.34 |
C9orf117 |
chromosome 9 open reading frame 117 |
7502 |
0.11 |
| chr11_806204_806355 | 0.34 |
PIDD |
p53-induced death domain protein |
1034 |
0.23 |
| chr10_33424940_33425136 | 0.34 |
ENSG00000263576 |
. |
37474 |
0.16 |
| chr5_81365043_81365194 | 0.34 |
ATG10-AS1 |
ATG10 antisense RNA 1 |
4403 |
0.24 |
| chr8_23075253_23075404 | 0.33 |
ENSG00000246582 |
. |
6656 |
0.12 |
| chr1_245316415_245317636 | 0.33 |
KIF26B |
kinesin family member 26B |
1262 |
0.47 |
| chr2_225823561_225823839 | 0.33 |
DOCK10 |
dedicator of cytokinesis 10 |
11918 |
0.28 |
| chr14_76298268_76298419 | 0.33 |
RP11-270M14.4 |
|
5211 |
0.29 |
| chr1_32393915_32394066 | 0.33 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
8906 |
0.18 |
| chr2_225750747_225750920 | 0.32 |
DOCK10 |
dedicator of cytokinesis 10 |
60949 |
0.15 |
| chr11_70244216_70245100 | 0.32 |
CTTN |
cortactin |
11 |
0.53 |
| chr13_46741429_46741580 | 0.32 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
1150 |
0.42 |
| chr5_102098307_102098531 | 0.32 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
7372 |
0.33 |
| chr1_109597558_109597709 | 0.32 |
WDR47 |
WD repeat domain 47 |
12783 |
0.12 |
| chr1_172748125_172748386 | 0.32 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
120097 |
0.06 |
| chr2_73496881_73497091 | 0.32 |
FBXO41 |
F-box protein 41 |
1057 |
0.43 |
| chr11_118478413_118479735 | 0.31 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
716 |
0.54 |
| chr4_127820346_127820497 | 0.31 |
ENSG00000199862 |
. |
128953 |
0.06 |
| chr1_145427603_145427863 | 0.31 |
TXNIP |
thioredoxin interacting protein |
10736 |
0.12 |
| chr9_117134390_117135168 | 0.31 |
AKNA |
AT-hook transcription factor |
4465 |
0.23 |
| chr9_128408983_128409218 | 0.31 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
3596 |
0.32 |
| chr3_5231863_5232390 | 0.31 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
2092 |
0.23 |
| chr6_160842707_160842858 | 0.31 |
SLC22A3 |
solute carrier family 22 (organic cation transporter), member 3 |
73357 |
0.11 |
| chr5_55786648_55786799 | 0.31 |
CTC-236F12.4 |
Uncharacterized protein |
9127 |
0.21 |
| chr1_25290384_25290732 | 0.31 |
RUNX3 |
runt-related transcription factor 3 |
917 |
0.57 |
| chr1_118403883_118404183 | 0.31 |
GDAP2 |
ganglioside induced differentiation associated protein 2 |
68173 |
0.12 |
| chr19_11160042_11160193 | 0.30 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
9394 |
0.14 |
| chr6_45837974_45838125 | 0.30 |
CLIC5 |
chloride intracellular channel 5 |
145516 |
0.04 |
| chr9_130614412_130614666 | 0.30 |
ENG |
endoglin |
2376 |
0.13 |
| chr7_73509063_73509569 | 0.30 |
LIMK1 |
LIM domain kinase 1 |
1907 |
0.33 |
| chr14_22309331_22309482 | 0.30 |
ENSG00000222776 |
. |
60621 |
0.14 |
| chr16_11676228_11676379 | 0.30 |
LITAF |
lipopolysaccharide-induced TNF factor |
3926 |
0.23 |
| chr19_14551433_14551660 | 0.30 |
PKN1 |
protein kinase N1 |
474 |
0.7 |
| chr9_81972295_81972446 | 0.30 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
214318 |
0.03 |
| chr7_90798408_90798559 | 0.30 |
FZD1 |
frizzled family receptor 1 |
95300 |
0.09 |
| chr18_2979843_2980201 | 0.30 |
LPIN2 |
lipin 2 |
2849 |
0.22 |
| chr20_20724641_20724860 | 0.30 |
ENSG00000264361 |
. |
5376 |
0.28 |
| chr9_116282321_116282472 | 0.30 |
RGS3 |
regulator of G-protein signaling 3 |
2846 |
0.31 |
| chr7_155326313_155326555 | 0.30 |
CNPY1 |
canopy FGF signaling regulator 1 |
99 |
0.97 |
| chr6_138248657_138248808 | 0.30 |
RP11-356I2.4 |
|
59362 |
0.13 |
| chr20_33871790_33873092 | 0.30 |
EIF6 |
eukaryotic translation initiation factor 6 |
77 |
0.88 |
| chrX_130036036_130037401 | 0.30 |
ENOX2 |
ecto-NOX disulfide-thiol exchanger 2 |
460 |
0.9 |
| chr5_89123597_89123748 | 0.30 |
ENSG00000264342 |
. |
188865 |
0.03 |
| chr9_89870217_89870368 | 0.30 |
ENSG00000212421 |
. |
5073 |
0.33 |
| chr17_73246169_73246352 | 0.30 |
GGA3 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 3 |
11098 |
0.09 |
| chrX_70766986_70767137 | 0.29 |
OGT |
O-linked N-acetylglucosamine (GlcNAc) transferase |
10197 |
0.18 |
| chr3_16342520_16342671 | 0.29 |
RP11-415F23.2 |
|
13351 |
0.16 |
| chr5_130602420_130602583 | 0.29 |
CDC42SE2 |
CDC42 small effector 2 |
2708 |
0.39 |
| chr2_12684277_12684428 | 0.29 |
ENSG00000207183 |
. |
132725 |
0.05 |
| chr15_78365295_78365472 | 0.29 |
TBC1D2B |
TBC1 domain family, member 2B |
4611 |
0.15 |
| chr10_12085799_12085950 | 0.29 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
851 |
0.6 |
| chr6_53198691_53198842 | 0.29 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
14821 |
0.2 |
| chr7_50361038_50361265 | 0.29 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
6094 |
0.31 |
| chr19_50836422_50837123 | 0.29 |
KCNC3 |
potassium voltage-gated channel, Shaw-related subfamily, member 3 |
0 |
0.95 |
| chr1_115605656_115605807 | 0.29 |
TSPAN2 |
tetraspanin 2 |
26304 |
0.2 |
| chr3_142227327_142227499 | 0.28 |
RP11-383G6.3 |
|
36234 |
0.13 |
| chr22_34318362_34318970 | 0.28 |
LARGE |
like-glycosyltransferase |
82 |
0.74 |
| chr10_63991622_63991773 | 0.28 |
RTKN2 |
rhotekin 2 |
4325 |
0.32 |
| chr2_55379810_55379961 | 0.28 |
RTN4 |
reticulon 4 |
40128 |
0.12 |
| chr18_27729065_27729216 | 0.28 |
ENSG00000221731 |
. |
665847 |
0.0 |
| chr15_55095080_55095231 | 0.28 |
UNC13C |
unc-13 homolog C (C. elegans) |
191482 |
0.03 |
| chr8_125615976_125616127 | 0.28 |
RP11-532M24.1 |
|
24179 |
0.17 |
| chr2_200899058_200899209 | 0.28 |
C2orf47 |
chromosome 2 open reading frame 47 |
78589 |
0.09 |
| chr5_57368587_57368738 | 0.28 |
ENSG00000238899 |
. |
2647 |
0.43 |
| chrX_24484452_24484603 | 0.28 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
954 |
0.63 |
| chr1_150484995_150485146 | 0.28 |
ECM1 |
extracellular matrix protein 1 |
4487 |
0.12 |
| chr11_96073413_96073903 | 0.28 |
ENSG00000266192 |
. |
944 |
0.55 |
| chr2_161993456_161993757 | 0.28 |
TANK |
TRAF family member-associated NFKB activator |
140 |
0.97 |
| chr11_62444789_62444940 | 0.28 |
UBXN1 |
UBX domain protein 1 |
591 |
0.42 |
| chr11_36641126_36641415 | 0.28 |
RAG2 |
recombination activating gene 2 |
21484 |
0.19 |
| chr1_29255082_29255406 | 0.28 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
14153 |
0.18 |
| chr17_27467519_27468040 | 0.28 |
RP11-321A17.4 |
|
278 |
0.55 |
| chr9_123606315_123606466 | 0.28 |
PSMD5-AS1 |
PSMD5 antisense RNA 1 (head to head) |
601 |
0.6 |
| chr5_75699014_75699893 | 0.28 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
304 |
0.94 |
| chr11_104951685_104951836 | 0.28 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
20398 |
0.15 |
| chr14_22565525_22565676 | 0.27 |
ENSG00000238634 |
. |
45287 |
0.18 |
| chr18_26676772_26676923 | 0.27 |
ENSG00000265730 |
. |
1204 |
0.6 |
| chr17_43486558_43486834 | 0.27 |
ARHGAP27 |
Rho GTPase activating protein 27 |
1056 |
0.39 |
| chr15_81591495_81592151 | 0.27 |
IL16 |
interleukin 16 |
66 |
0.98 |
| chr19_42391098_42391303 | 0.27 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
2685 |
0.17 |
| chr1_27945148_27945299 | 0.27 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
5350 |
0.15 |
| chr4_17205389_17205540 | 0.27 |
ENSG00000206780 |
. |
117041 |
0.07 |
| chr2_175462178_175462605 | 0.27 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
102 |
0.97 |
| chr8_83009707_83009858 | 0.27 |
SNX16 |
sorting nexin 16 |
254681 |
0.02 |
| chr8_125617266_125617519 | 0.27 |
RP11-532M24.1 |
|
25520 |
0.16 |
| chr5_14268280_14268580 | 0.27 |
TRIO |
trio Rho guanine nucleotide exchange factor |
22656 |
0.28 |
| chr16_3452838_3453133 | 0.27 |
ZNF174 |
zinc finger protein 174 |
1011 |
0.36 |
| chr9_130855530_130855681 | 0.27 |
RP11-379C10.4 |
|
1131 |
0.27 |
| chr7_94039789_94039940 | 0.27 |
COL1A2 |
collagen, type I, alpha 2 |
15991 |
0.25 |
| chr3_32995541_32995692 | 0.27 |
CCR4 |
chemokine (C-C motif) receptor 4 |
2550 |
0.37 |
| chr17_37931079_37931230 | 0.27 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
3324 |
0.18 |
| chr16_1730441_1730592 | 0.26 |
HN1L |
hematological and neurological expressed 1-like |
178 |
0.89 |
| chr20_62258641_62259758 | 0.26 |
CTD-3184A7.4 |
|
596 |
0.45 |
| chrX_19815653_19816006 | 0.26 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
2040 |
0.46 |
| chr6_109262468_109262619 | 0.26 |
ARMC2-AS1 |
ARMC2 antisense RNA 1 |
17237 |
0.2 |
| chr3_115596546_115596697 | 0.26 |
ENSG00000243359 |
. |
39906 |
0.22 |
| chr4_154439708_154439859 | 0.26 |
KIAA0922 |
KIAA0922 |
37265 |
0.19 |
| chr2_122660445_122660607 | 0.26 |
TSN |
translin |
146779 |
0.04 |
| chr2_182322868_182323440 | 0.26 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
999 |
0.69 |
| chrX_78201853_78202096 | 0.26 |
P2RY10 |
purinergic receptor P2Y, G-protein coupled, 10 |
1056 |
0.69 |
| chr17_56416209_56416360 | 0.26 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
1565 |
0.24 |
| chr17_74235344_74235495 | 0.26 |
RNF157 |
ring finger protein 157 |
963 |
0.47 |
| chr3_187005158_187005309 | 0.26 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
4252 |
0.27 |
| chr13_52397484_52397635 | 0.26 |
RP11-327P2.5 |
|
19126 |
0.17 |
| chr1_226922484_226922931 | 0.26 |
ITPKB |
inositol-trisphosphate 3-kinase B |
2452 |
0.34 |
| chr4_41122983_41123211 | 0.26 |
ENSG00000207198 |
. |
7138 |
0.23 |
| chr17_42072137_42072373 | 0.26 |
PYY |
peptide YY |
9582 |
0.1 |
| chr2_143905734_143905885 | 0.25 |
ARHGAP15 |
Rho GTPase activating protein 15 |
18926 |
0.23 |
| chr4_7878163_7878536 | 0.25 |
AFAP1 |
actin filament associated protein 1 |
4309 |
0.28 |
| chr1_155900220_155900371 | 0.25 |
KIAA0907 |
KIAA0907 |
3869 |
0.1 |
| chr10_92434851_92435027 | 0.25 |
HTR7 |
5-hydroxytryptamine (serotonin) receptor 7, adenylate cyclase-coupled |
182516 |
0.03 |
| chr6_26358532_26358683 | 0.25 |
ENSG00000252399 |
. |
5343 |
0.1 |
| chr3_56833713_56834040 | 0.25 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
2101 |
0.44 |
| chr1_9777067_9777879 | 0.25 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
654 |
0.69 |
| chr15_53096656_53097309 | 0.25 |
ONECUT1 |
one cut homeobox 1 |
14773 |
0.27 |
| chr2_231200345_231200592 | 0.25 |
SP140L |
SP140 nuclear body protein-like |
6317 |
0.26 |
| chr1_167484289_167484555 | 0.25 |
CD247 |
CD247 molecule |
3353 |
0.26 |
| chr9_139437724_139437875 | 0.25 |
RP11-413M3.4 |
|
466 |
0.63 |
| chr6_37017654_37018643 | 0.25 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
5541 |
0.22 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.6 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.2 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.2 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.1 | 0.3 | GO:0002664 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.3 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0002921 | negative regulation of humoral immune response(GO:0002921) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.2 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.2 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.6 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.1 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.2 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.2 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.2 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.1 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0071636 | positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.0 | 0.1 | GO:0046666 | retinal cell programmed cell death(GO:0046666) |
| 0.0 | 0.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.2 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.1 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.2 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.0 | GO:0060253 | negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.0 | GO:0060430 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.2 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0032071 | regulation of deoxyribonuclease activity(GO:0032070) regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:2001259 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0048541 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) regulation of gamma-delta T cell activation(GO:0046643) positive regulation of gamma-delta T cell activation(GO:0046645) Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.2 | GO:0071384 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
| 0.0 | 0.1 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.0 | GO:0072239 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.0 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.0 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.2 | GO:0042308 | negative regulation of protein import into nucleus(GO:0042308) negative regulation of protein localization to nucleus(GO:1900181) negative regulation of protein import(GO:1904590) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0070170 | regulation of tooth mineralization(GO:0070170) |
| 0.0 | 0.1 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.0 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.0 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) regulation of signal transduction by p53 class mediator(GO:1901796) |
| 0.0 | 0.0 | GO:0042940 | D-amino acid transport(GO:0042940) D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) auditory receptor cell fate determination(GO:0042668) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.2 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.2 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:0019079 | viral genome replication(GO:0019079) |
| 0.0 | 0.5 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.3 | GO:0051017 | actin filament bundle assembly(GO:0051017) actin filament bundle organization(GO:0061572) |
| 0.0 | 0.1 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.0 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.6 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.1 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.3 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.2 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.5 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0001640 | adenylate cyclase inhibiting G-protein coupled glutamate receptor activity(GO:0001640) G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.0 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.5 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.0 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.1 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.8 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.0 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.4 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.0 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.0 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.0 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |