Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
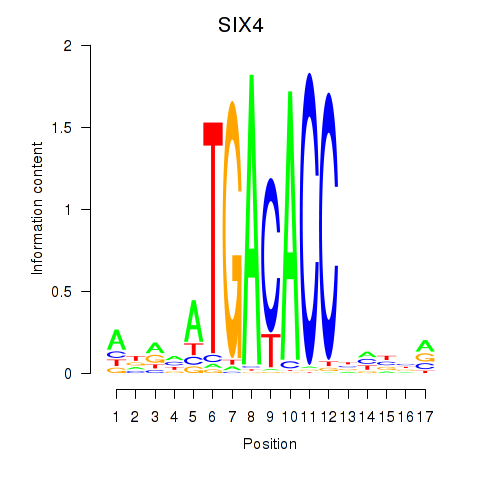
Results for SIX4
Z-value: 0.52
Transcription factors associated with SIX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SIX4
|
ENSG00000100625.8 | SIX homeobox 4 |
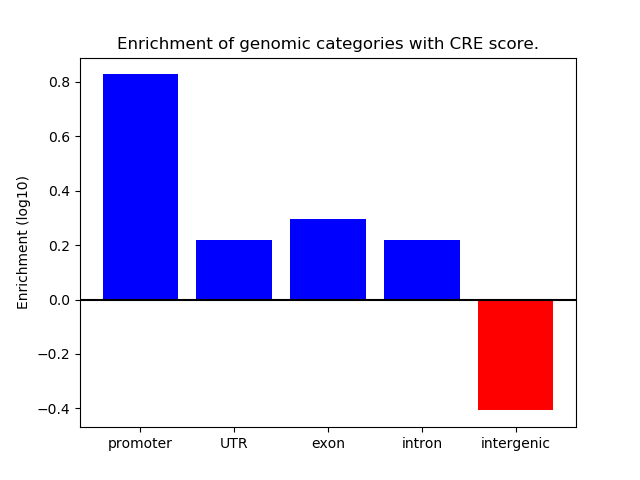
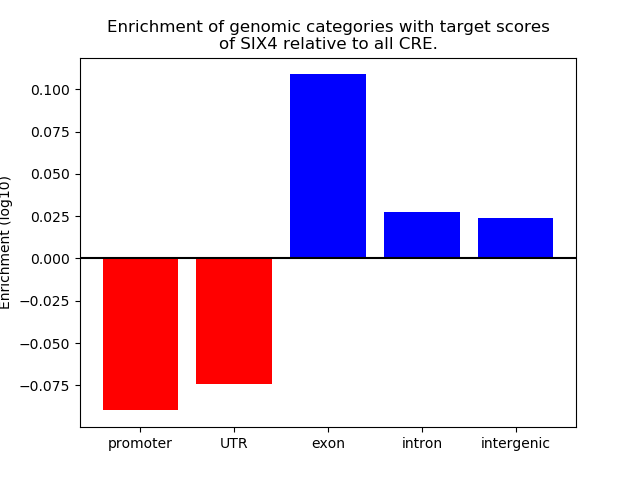
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_61190612_61190839 | SIX4 | 127 | 0.967784 | -0.35 | 3.6e-01 | Click! |
| chr14_61188583_61188797 | SIX4 | 2162 | 0.334416 | -0.30 | 4.4e-01 | Click! |
| chr14_61188418_61188569 | SIX4 | 2359 | 0.316607 | -0.25 | 5.2e-01 | Click! |
| chr14_61190877_61191072 | SIX4 | 92 | 0.973959 | -0.24 | 5.4e-01 | Click! |
| chr14_61188910_61189596 | SIX4 | 1599 | 0.413092 | -0.15 | 6.9e-01 | Click! |
Activity of the SIX4 motif across conditions
Conditions sorted by the z-value of the SIX4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
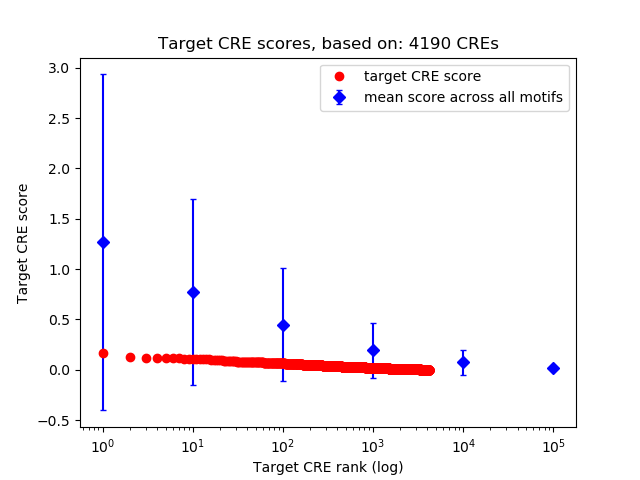
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_126308274_126308480 | 0.16 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
83817 |
0.08 |
| chr16_83620596_83620747 | 0.12 |
ENSG00000263785 |
. |
78720 |
0.09 |
| chr12_32050718_32050938 | 0.12 |
ENSG00000252584 |
. |
17813 |
0.2 |
| chr2_64894623_64894774 | 0.12 |
SERTAD2 |
SERTA domain containing 2 |
13651 |
0.23 |
| chr2_172734713_172734864 | 0.11 |
SLC25A12 |
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
15945 |
0.18 |
| chr22_19950191_19950342 | 0.11 |
COMT |
catechol-O-methyltransferase |
196 |
0.86 |
| chr8_53063329_53063558 | 0.11 |
RP11-26M5.3 |
|
63 |
0.98 |
| chr17_40937029_40937180 | 0.11 |
WNK4 |
WNK lysine deficient protein kinase 4 |
4408 |
0.09 |
| chr11_35949210_35949523 | 0.11 |
RP11-698N11.2 |
|
9077 |
0.19 |
| chr20_50664659_50664866 | 0.11 |
ZFP64 |
ZFP64 zinc finger protein |
50243 |
0.17 |
| chr11_100555302_100555453 | 0.11 |
ARHGAP42 |
Rho GTPase activating protein 42 |
3007 |
0.26 |
| chr4_165040749_165040900 | 0.11 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
264317 |
0.02 |
| chr12_80079262_80079413 | 0.10 |
PAWR |
PRKC, apoptosis, WT1, regulator |
4523 |
0.3 |
| chr12_107631394_107631672 | 0.10 |
BTBD11 |
BTB (POZ) domain containing 11 |
80657 |
0.1 |
| chr7_107587877_107588028 | 0.10 |
CTB-13F3.1 |
|
5391 |
0.21 |
| chr7_73682843_73682994 | 0.10 |
ENSG00000252538 |
. |
9972 |
0.16 |
| chr7_964241_964392 | 0.10 |
ADAP1 |
ArfGAP with dual PH domains 1 |
2536 |
0.22 |
| chr2_196434123_196434274 | 0.10 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
6503 |
0.28 |
| chr12_123368397_123368548 | 0.10 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
6234 |
0.18 |
| chr22_28063217_28063368 | 0.10 |
RP11-375H17.1 |
|
49176 |
0.18 |
| chr6_46702576_46703631 | 0.09 |
PLA2G7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
24 |
0.97 |
| chr18_25667459_25667610 | 0.09 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
9659 |
0.32 |
| chr1_957490_957641 | 0.09 |
AGRN |
agrin |
2062 |
0.15 |
| chr20_49378517_49378668 | 0.09 |
PARD6B |
par-6 family cell polarity regulator beta |
30390 |
0.14 |
| chr3_5857555_5857706 | 0.09 |
ENSG00000241227 |
. |
562722 |
0.0 |
| chr21_33744937_33745088 | 0.09 |
ENSG00000200792 |
. |
4619 |
0.15 |
| chr2_161094906_161095057 | 0.09 |
ITGB6 |
integrin, beta 6 |
38166 |
0.21 |
| chr13_101262503_101262654 | 0.09 |
GGACT |
gamma-glutamylamine cyclotransferase |
20796 |
0.19 |
| chr2_75043587_75043738 | 0.09 |
HK2 |
hexokinase 2 |
17446 |
0.23 |
| chr2_38265076_38265999 | 0.08 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
2053 |
0.33 |
| chr10_28378559_28378902 | 0.08 |
ARMC4 |
armadillo repeat containing 4 |
90753 |
0.09 |
| chr6_155438873_155439024 | 0.08 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
4102 |
0.33 |
| chr8_104384027_104384290 | 0.08 |
CTHRC1 |
collagen triple helix repeat containing 1 |
380 |
0.83 |
| chr14_75689238_75689389 | 0.08 |
ENSG00000252013 |
. |
20441 |
0.14 |
| chr14_22749370_22749521 | 0.08 |
ENSG00000238634 |
. |
138558 |
0.04 |
| chr1_204333064_204333215 | 0.08 |
PLEKHA6 |
pleckstrin homology domain containing, family A member 6 |
4095 |
0.21 |
| chr11_3923942_3924093 | 0.08 |
STIM1 |
stromal interaction molecule 1 |
676 |
0.62 |
| chr16_13520212_13520363 | 0.08 |
AC003009.1 |
|
93847 |
0.09 |
| chr17_78912361_78912527 | 0.08 |
CTD-2561B21.4 |
|
3389 |
0.17 |
| chr20_31888809_31889213 | 0.08 |
BPIFB1 |
BPI fold containing family B, member 1 |
18070 |
0.15 |
| chr16_66008915_66009066 | 0.08 |
ENSG00000201999 |
. |
326722 |
0.01 |
| chr20_50104056_50104339 | 0.08 |
ENSG00000266761 |
. |
34683 |
0.19 |
| chr19_2089521_2089711 | 0.08 |
MOB3A |
MOB kinase activator 3A |
559 |
0.63 |
| chr1_245316415_245317636 | 0.08 |
KIF26B |
kinesin family member 26B |
1262 |
0.47 |
| chr16_23084021_23084263 | 0.08 |
USP31 |
ubiquitin specific peptidase 31 |
273 |
0.95 |
| chr6_168473471_168473622 | 0.08 |
FRMD1 |
FERM domain containing 1 |
3000 |
0.33 |
| chr22_25346267_25346418 | 0.08 |
KIAA1671 |
KIAA1671 |
2355 |
0.29 |
| chr11_133459496_133459647 | 0.08 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
57157 |
0.15 |
| chr1_214499564_214499715 | 0.07 |
SMYD2 |
SET and MYND domain containing 2 |
4681 |
0.29 |
| chr1_109597558_109597709 | 0.07 |
WDR47 |
WD repeat domain 47 |
12783 |
0.12 |
| chr11_117881316_117881467 | 0.07 |
IL10RA |
interleukin 10 receptor, alpha |
24282 |
0.15 |
| chr1_43411974_43412125 | 0.07 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
12451 |
0.18 |
| chr12_66122461_66123138 | 0.07 |
HMGA2 |
high mobility group AT-hook 2 |
95112 |
0.08 |
| chr3_165828020_165828171 | 0.07 |
BCHE |
butyrylcholinesterase |
272835 |
0.02 |
| chr15_48937037_48938293 | 0.07 |
FBN1 |
fibrillin 1 |
253 |
0.95 |
| chr7_118512519_118512670 | 0.07 |
ENSG00000222226 |
. |
410041 |
0.01 |
| chr3_195713973_195714124 | 0.07 |
SDHAP1 |
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 1 |
3081 |
0.23 |
| chr1_9048089_9048240 | 0.07 |
ENSG00000265141 |
. |
8231 |
0.17 |
| chr2_238358341_238358492 | 0.07 |
AC112721.1 |
Uncharacterized protein |
24497 |
0.16 |
| chr7_140677362_140677513 | 0.07 |
MRPS33 |
mitochondrial ribosomal protein S33 |
36792 |
0.15 |
| chr2_70750715_70751050 | 0.07 |
TGFA |
transforming growth factor, alpha |
29740 |
0.17 |
| chr3_184037306_184037783 | 0.07 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
1 |
0.95 |
| chr20_1602572_1602723 | 0.07 |
RP4-576H24.4 |
Uncharacterized protein |
1992 |
0.21 |
| chr20_11098246_11098397 | 0.07 |
C20orf187 |
chromosome 20 open reading frame 187 |
89510 |
0.1 |
| chr1_95388573_95389173 | 0.07 |
CNN3 |
calponin 3, acidic |
2464 |
0.27 |
| chr3_157924014_157924165 | 0.07 |
RSRC1 |
arginine/serine-rich coiled-coil 1 |
3228 |
0.38 |
| chr4_3391445_3391596 | 0.07 |
RGS12 |
regulator of G-protein signaling 12 |
3459 |
0.27 |
| chr17_16177405_16177556 | 0.07 |
ENSG00000221355 |
. |
7848 |
0.18 |
| chr6_53458265_53458416 | 0.07 |
GCLC |
glutamate-cysteine ligase, catalytic subunit |
23428 |
0.17 |
| chr2_27890028_27890179 | 0.07 |
SUPT7L |
suppressor of Ty 7 (S. cerevisiae)-like |
3427 |
0.14 |
| chr2_216288358_216288696 | 0.07 |
FN1 |
fibronectin 1 |
12263 |
0.21 |
| chrX_111124795_111124946 | 0.07 |
TRPC5OS |
TRPC5 opposite strand |
255 |
0.96 |
| chr20_52324568_52324793 | 0.07 |
ENSG00000238468 |
. |
39383 |
0.19 |
| chr3_134036563_134036714 | 0.07 |
AMOTL2 |
angiomotin like 2 |
54116 |
0.13 |
| chr7_73304367_73304518 | 0.07 |
WBSCR28 |
Williams-Beuren syndrome chromosome region 28 |
28953 |
0.15 |
| chr15_71606160_71606311 | 0.07 |
RP11-592N21.2 |
|
28428 |
0.23 |
| chr20_4142074_4142225 | 0.07 |
SMOX |
spermine oxidase |
10208 |
0.21 |
| chr3_99791321_99791472 | 0.07 |
FILIP1L |
filamin A interacting protein 1-like |
41961 |
0.15 |
| chr1_77942138_77942289 | 0.07 |
AK5 |
adenylate kinase 5 |
55579 |
0.12 |
| chr22_38199332_38199483 | 0.07 |
H1F0 |
H1 histone family, member 0 |
1707 |
0.2 |
| chr9_133838398_133838632 | 0.07 |
FIBCD1 |
fibrinogen C domain containing 1 |
23988 |
0.14 |
| chr17_19769997_19771171 | 0.07 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
646 |
0.74 |
| chr6_159674242_159674415 | 0.07 |
FNDC1-IT1 |
FNDC1 intronic transcript 1 (non-protein coding) |
12510 |
0.24 |
| chr2_136577729_136578391 | 0.07 |
AC011893.3 |
|
299 |
0.89 |
| chr1_180062348_180062499 | 0.07 |
CEP350 |
centrosomal protein 350kDa |
426 |
0.88 |
| chr1_244264308_244264663 | 0.07 |
ENSG00000244066 |
. |
2749 |
0.32 |
| chr7_107045766_107045917 | 0.07 |
GPR22 |
G protein-coupled receptor 22 |
64664 |
0.11 |
| chr5_58858854_58859005 | 0.07 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
23290 |
0.28 |
| chr22_30658388_30658630 | 0.07 |
OSM |
oncostatin M |
3298 |
0.13 |
| chr4_129732983_129734245 | 0.07 |
JADE1 |
jade family PHD finger 1 |
615 |
0.83 |
| chr16_23515749_23516009 | 0.07 |
GGA2 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
5831 |
0.15 |
| chr7_55620313_55620510 | 0.07 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
134 |
0.97 |
| chr11_68049808_68050049 | 0.06 |
C11orf24 |
chromosome 11 open reading frame 24 |
10471 |
0.2 |
| chr2_79951275_79951426 | 0.06 |
ENSG00000263566 |
. |
70046 |
0.1 |
| chr16_53086342_53086851 | 0.06 |
RP11-467J12.4 |
|
189 |
0.95 |
| chr13_67597561_67597712 | 0.06 |
PCDH9-AS4 |
PCDH9 antisense RNA 4 |
32618 |
0.21 |
| chr4_154412529_154412680 | 0.06 |
KIAA0922 |
KIAA0922 |
25103 |
0.22 |
| chr3_49137947_49138098 | 0.06 |
QARS |
glutaminyl-tRNA synthetase |
994 |
0.34 |
| chr9_136009638_136009789 | 0.06 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
3209 |
0.19 |
| chr2_6221801_6221952 | 0.06 |
DKFZP761K2322 |
HCG1990367; Putative uncharacterized protein DKFZp761K2322; Putative uncharacterized protein FLJ30594; Uncharacterized protein; cDNA FLJ30594 fis, clone BRAWH2008903 |
110164 |
0.08 |
| chr16_9204517_9204668 | 0.06 |
RP11-473I1.10 |
|
3494 |
0.19 |
| chr5_73929379_73929530 | 0.06 |
HEXB |
hexosaminidase B (beta polypeptide) |
6394 |
0.22 |
| chr20_57235059_57235243 | 0.06 |
STX16 |
syntaxin 16 |
7425 |
0.17 |
| chr2_50719361_50719512 | 0.06 |
NRXN1 |
neurexin 1 |
3778 |
0.38 |
| chr7_28530451_28530602 | 0.06 |
CREB5 |
cAMP responsive element binding protein 5 |
222 |
0.97 |
| chr5_17400004_17400276 | 0.06 |
ENSG00000201715 |
. |
54415 |
0.16 |
| chr14_70117061_70117212 | 0.06 |
KIAA0247 |
KIAA0247 |
38823 |
0.17 |
| chr2_198077334_198077485 | 0.06 |
ANKRD44 |
ankyrin repeat domain 44 |
14647 |
0.2 |
| chr2_28601499_28601815 | 0.06 |
FOSL2 |
FOS-like antigen 2 |
14012 |
0.17 |
| chr22_29601932_29602692 | 0.06 |
EMID1 |
EMI domain containing 1 |
400 |
0.8 |
| chr16_28158064_28158215 | 0.06 |
XPO6 |
exportin 6 |
23083 |
0.16 |
| chr12_122460681_122461523 | 0.06 |
BCL7A |
B-cell CLL/lymphoma 7A |
1310 |
0.49 |
| chr18_45463703_45463854 | 0.06 |
SMAD2 |
SMAD family member 2 |
6263 |
0.28 |
| chr1_210465594_210466382 | 0.06 |
HHAT |
hedgehog acyltransferase |
35608 |
0.17 |
| chr2_174436276_174436427 | 0.06 |
CDCA7 |
cell division cycle associated 7 |
216751 |
0.02 |
| chr2_235397051_235397202 | 0.06 |
ARL4C |
ADP-ribosylation factor-like 4C |
8118 |
0.33 |
| chr10_11192421_11192572 | 0.06 |
CELF2 |
CUGBP, Elav-like family member 2 |
14497 |
0.2 |
| chr4_185827450_185827601 | 0.06 |
ENSG00000263458 |
. |
32069 |
0.17 |
| chr15_84033783_84034563 | 0.06 |
BNC1 |
basonuclin 1 |
80707 |
0.09 |
| chr12_27742378_27742529 | 0.06 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
9828 |
0.23 |
| chr3_46994537_46994688 | 0.06 |
CCDC12 |
coiled-coil domain containing 12 |
23658 |
0.14 |
| chr8_48515171_48515322 | 0.06 |
SPIDR |
scaffolding protein involved in DNA repair |
56980 |
0.12 |
| chr21_44036406_44036646 | 0.06 |
AP001626.2 |
|
34828 |
0.13 |
| chr21_30720865_30721016 | 0.06 |
BACH1-IT1 |
BACH1 intronic transcript 1 (non-protein coding) |
3015 |
0.22 |
| chrX_19716745_19716896 | 0.06 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
27704 |
0.25 |
| chr22_45642723_45642874 | 0.06 |
KIAA0930 |
KIAA0930 |
6148 |
0.19 |
| chr12_65032414_65032565 | 0.06 |
RP11-338E21.2 |
|
10365 |
0.12 |
| chr15_68574240_68574639 | 0.06 |
FEM1B |
fem-1 homolog b (C. elegans) |
2142 |
0.31 |
| chr5_6349990_6350141 | 0.06 |
MED10 |
mediator complex subunit 10 |
28642 |
0.24 |
| chr16_21518478_21518933 | 0.06 |
ENSG00000265462 |
. |
1249 |
0.41 |
| chr3_31719857_31720008 | 0.06 |
OSBPL10-AS1 |
OSBPL10 antisense RNA 1 |
25618 |
0.25 |
| chr7_44922298_44922675 | 0.06 |
ENSG00000264326 |
. |
1087 |
0.34 |
| chr8_128911768_128911919 | 0.06 |
TMEM75 |
transmembrane protein 75 |
48748 |
0.14 |
| chrX_144503620_144503771 | 0.06 |
SPANXN1 |
SPANX family, member N1 |
175347 |
0.03 |
| chr4_148675376_148675527 | 0.06 |
ENSG00000252951 |
. |
8883 |
0.18 |
| chr4_7972052_7972961 | 0.06 |
AFAP1 |
actin filament associated protein 1 |
30853 |
0.14 |
| chr15_99386539_99386690 | 0.06 |
IGF1R |
insulin-like growth factor 1 receptor |
46956 |
0.14 |
| chr10_131754567_131754718 | 0.06 |
EBF3 |
early B-cell factor 3 |
7463 |
0.31 |
| chr18_47090550_47090839 | 0.06 |
LIPG |
lipase, endothelial |
1979 |
0.29 |
| chr11_79833397_79833684 | 0.06 |
ENSG00000200146 |
. |
405464 |
0.01 |
| chr11_124940847_124941136 | 0.06 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
7759 |
0.19 |
| chr3_151744346_151744509 | 0.06 |
RP11-454C18.2 |
|
98464 |
0.08 |
| chr12_22781335_22781486 | 0.06 |
ETNK1 |
ethanolamine kinase 1 |
3119 |
0.28 |
| chr3_179191370_179191536 | 0.06 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
22075 |
0.17 |
| chr7_96646152_96646303 | 0.06 |
DLX6-AS1 |
DLX6 antisense RNA 1 |
2850 |
0.21 |
| chr15_68958574_68958725 | 0.06 |
CORO2B |
coronin, actin binding protein, 2B |
34322 |
0.21 |
| chr12_95609361_95609804 | 0.06 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
1573 |
0.32 |
| chr17_41608232_41608498 | 0.05 |
ETV4 |
ets variant 4 |
563 |
0.68 |
| chr8_134261990_134262245 | 0.05 |
WISP1-OT1 |
WISP1 overlapping transcript 1 (non-protein coding) |
20818 |
0.21 |
| chr10_48600586_48600737 | 0.05 |
GDF10 |
growth differentiation factor 10 |
161685 |
0.03 |
| chr6_155599532_155599683 | 0.05 |
CLDN20 |
claudin 20 |
14460 |
0.19 |
| chr18_54741654_54741805 | 0.05 |
WDR7-OT1 |
WDR7 overlapping transcript 1 (non-protein coding) |
46666 |
0.16 |
| chr4_15685369_15685921 | 0.05 |
FAM200B |
family with sequence similarity 200, member B |
2216 |
0.26 |
| chr1_234792015_234792239 | 0.05 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
46856 |
0.14 |
| chr3_190038156_190038307 | 0.05 |
CLDN1 |
claudin 1 |
2033 |
0.41 |
| chr17_39739860_39740150 | 0.05 |
KRT14 |
keratin 14 |
3168 |
0.13 |
| chr12_124386990_124387141 | 0.05 |
DNAH10 |
dynein, axonemal, heavy chain 10 |
5795 |
0.12 |
| chr15_74217167_74217391 | 0.05 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
1360 |
0.3 |
| chr12_94634224_94634375 | 0.05 |
PLXNC1 |
plexin C1 |
13995 |
0.18 |
| chr5_141977695_141977846 | 0.05 |
AC005592.2 |
|
16357 |
0.19 |
| chr6_21264625_21264776 | 0.05 |
ENSG00000252046 |
. |
270901 |
0.02 |
| chr6_154735371_154735522 | 0.05 |
CNKSR3 |
CNKSR family member 3 |
16183 |
0.26 |
| chr18_71356092_71356243 | 0.05 |
ENSG00000241866 |
. |
56003 |
0.18 |
| chr7_80412610_80412761 | 0.05 |
CD36 |
CD36 molecule (thrombospondin receptor) |
109993 |
0.07 |
| chr9_90356869_90357020 | 0.05 |
ENSG00000222408 |
. |
2964 |
0.25 |
| chr15_84363051_84363202 | 0.05 |
ENSG00000212374 |
. |
23053 |
0.2 |
| chr12_16501163_16501421 | 0.05 |
MGST1 |
microsomal glutathione S-transferase 1 |
561 |
0.83 |
| chr2_16616720_16616871 | 0.05 |
AC104623.2 |
|
87649 |
0.1 |
| chr4_186227680_186227831 | 0.05 |
RP11-714G18.1 |
|
64124 |
0.08 |
| chr14_22538566_22539116 | 0.05 |
ENSG00000238634 |
. |
72046 |
0.12 |
| chr17_17641829_17642128 | 0.05 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
32157 |
0.12 |
| chr2_173258453_173258604 | 0.05 |
ITGA6 |
integrin, alpha 6 |
33554 |
0.16 |
| chr4_77932231_77932404 | 0.05 |
SEPT11 |
septin 11 |
9368 |
0.24 |
| chr4_29417750_29417901 | 0.05 |
ENSG00000252303 |
. |
94432 |
0.1 |
| chr16_2656430_2656581 | 0.05 |
AC141586.5 |
|
3030 |
0.11 |
| chr20_1518089_1518396 | 0.05 |
ENSG00000252103 |
. |
8437 |
0.14 |
| chr4_148518499_148518650 | 0.05 |
RP11-425A23.1 |
|
19822 |
0.19 |
| chrX_111125134_111125285 | 0.05 |
TRPC5OS |
TRPC5 opposite strand |
84 |
0.98 |
| chr11_61739742_61739933 | 0.05 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
4384 |
0.15 |
| chr6_138249034_138249358 | 0.05 |
RP11-356I2.4 |
|
59826 |
0.13 |
| chr10_70847938_70848765 | 0.05 |
SRGN |
serglycin |
477 |
0.82 |
| chr16_80793490_80793641 | 0.05 |
CTD-2055G21.1 |
|
23308 |
0.19 |
| chr9_86451900_86452137 | 0.05 |
RP11-575L7.2 |
|
51 |
0.97 |
| chr20_33871790_33873092 | 0.05 |
EIF6 |
eukaryotic translation initiation factor 6 |
77 |
0.88 |
| chr11_108408309_108408460 | 0.05 |
EXPH5 |
exophilin 5 |
550 |
0.8 |
| chr3_171594335_171594540 | 0.05 |
TMEM212-IT1 |
TMEM212 intronic transcript 1 (non-protein coding) |
17789 |
0.2 |
| chr11_85894364_85894515 | 0.05 |
ENSG00000200877 |
. |
30170 |
0.18 |
| chr6_34632212_34632529 | 0.05 |
C6orf106 |
chromosome 6 open reading frame 106 |
7363 |
0.14 |
| chr21_40378453_40378604 | 0.05 |
ENSG00000272015 |
. |
111819 |
0.06 |
| chr9_14170879_14171030 | 0.05 |
NFIB |
nuclear factor I/B |
9837 |
0.32 |
| chr6_138070872_138071097 | 0.05 |
ENSG00000216097 |
. |
32899 |
0.17 |
| chr7_130132004_130132893 | 0.05 |
MEST |
mesoderm specific transcript |
135 |
0.93 |
| chr17_30114926_30115210 | 0.05 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
70882 |
0.09 |
| chr16_10934760_10934911 | 0.05 |
TVP23A |
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
22193 |
0.15 |
| chr1_2985287_2985858 | 0.05 |
PRDM16 |
PR domain containing 16 |
160 |
0.85 |
| chr5_137348056_137348207 | 0.05 |
RP11-325L7.1 |
|
20332 |
0.13 |
| chr2_173145916_173146187 | 0.05 |
ENSG00000238572 |
. |
125203 |
0.05 |
| chr7_28747986_28748200 | 0.05 |
CREB5 |
cAMP responsive element binding protein 5 |
22495 |
0.28 |
| chr15_49098614_49098784 | 0.05 |
CEP152 |
centrosomal protein 152kDa |
4092 |
0.19 |
| chr9_20377607_20377758 | 0.05 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
4785 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.0 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |