Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
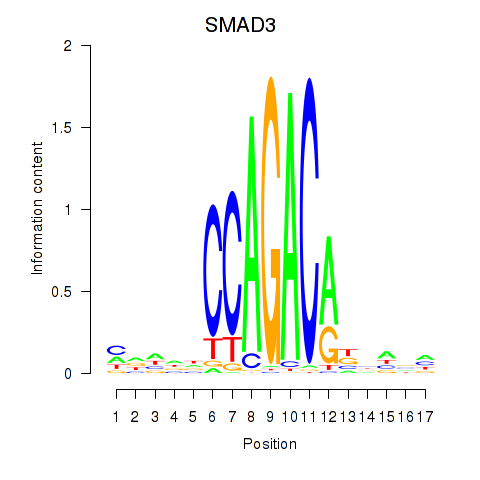
Results for SMAD3
Z-value: 0.73
Transcription factors associated with SMAD3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SMAD3
|
ENSG00000166949.11 | SMAD family member 3 |
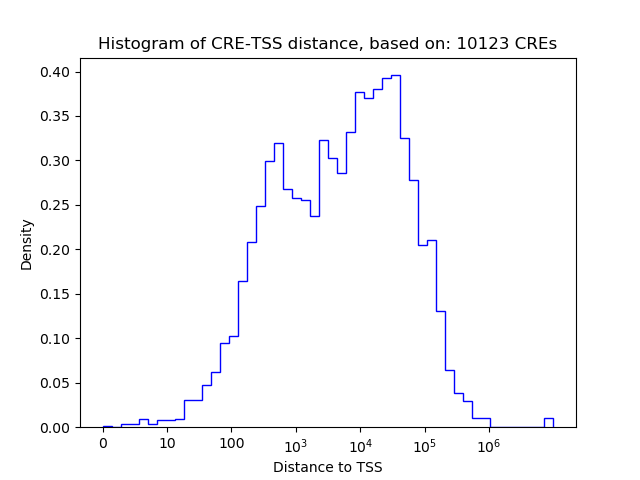
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_67311354_67311505 | SMAD3 | 44672 | 0.186105 | -0.87 | 2.5e-03 | Click! |
| chr15_67467161_67467312 | SMAD3 | 8367 | 0.225281 | -0.85 | 3.5e-03 | Click! |
| chr15_67413509_67413942 | SMAD3 | 4329 | 0.275328 | -0.81 | 8.3e-03 | Click! |
| chr15_67397321_67397472 | SMAD3 | 6379 | 0.268255 | -0.80 | 9.6e-03 | Click! |
| chr15_67394209_67394360 | SMAD3 | 3267 | 0.324710 | -0.77 | 1.6e-02 | Click! |
Activity of the SMAD3 motif across conditions
Conditions sorted by the z-value of the SMAD3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
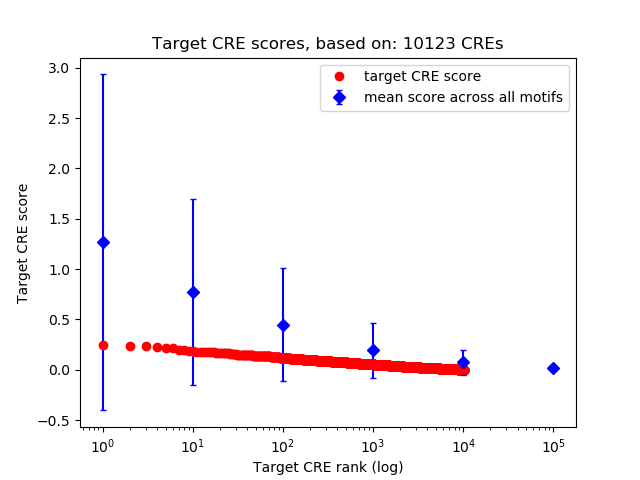
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_118481086_118481679 | 0.25 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
3024 |
0.16 |
| chr10_128250315_128250594 | 0.24 |
ENSG00000221717 |
. |
4821 |
0.29 |
| chr13_27927814_27927965 | 0.24 |
ENSG00000252247 |
. |
10006 |
0.16 |
| chr7_40510514_40510679 | 0.23 |
AC004988.1 |
|
75931 |
0.12 |
| chr2_149574916_149575083 | 0.22 |
ENSG00000222126 |
. |
8405 |
0.24 |
| chr1_110455050_110455346 | 0.21 |
CSF1 |
colony stimulating factor 1 (macrophage) |
1590 |
0.37 |
| chr11_44008945_44009331 | 0.20 |
C11orf96 |
chromosome 11 open reading frame 96 |
45083 |
0.11 |
| chr17_77767116_77767731 | 0.19 |
CBX8 |
chromobox homolog 8 |
3492 |
0.16 |
| chr1_159823893_159825316 | 0.19 |
C1orf204 |
chromosome 1 open reading frame 204 |
533 |
0.6 |
| chr7_100200060_100200259 | 0.18 |
PCOLCE |
procollagen C-endopeptidase enhancer |
359 |
0.52 |
| chrX_106873865_106874016 | 0.18 |
PRPS1 |
phosphoribosyl pyrophosphate synthetase 1 |
2147 |
0.39 |
| chr16_81551895_81552046 | 0.18 |
CMIP |
c-Maf inducing protein |
23016 |
0.25 |
| chr16_82769509_82769673 | 0.18 |
RP11-22H5.2 |
|
37333 |
0.19 |
| chr2_191915750_191915901 | 0.18 |
ENSG00000231858 |
. |
29573 |
0.14 |
| chr6_45705656_45705807 | 0.17 |
ENSG00000252738 |
. |
91890 |
0.09 |
| chr2_106712942_106713243 | 0.17 |
C2orf40 |
chromosome 2 open reading frame 40 |
30836 |
0.19 |
| chr2_42275788_42276453 | 0.17 |
PKDCC |
protein kinase domain containing, cytoplasmic |
960 |
0.61 |
| chr5_143171507_143171658 | 0.17 |
HMHB1 |
histocompatibility (minor) HB-1 |
20144 |
0.27 |
| chr5_15501266_15501663 | 0.17 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
83 |
0.99 |
| chr4_148471587_148471738 | 0.17 |
EDNRA |
endothelin receptor type A |
64828 |
0.11 |
| chr5_154832195_154832346 | 0.17 |
ENSG00000221131 |
. |
427910 |
0.01 |
| chr10_113968621_113968821 | 0.17 |
GPAM |
glycerol-3-phosphate acyltransferase, mitochondrial |
25196 |
0.23 |
| chrX_57162974_57163409 | 0.17 |
SPIN2A |
spindlin family, member 2A |
239 |
0.93 |
| chr12_50067665_50067882 | 0.17 |
FMNL3 |
formin-like 3 |
33230 |
0.11 |
| chr4_96336664_96336815 | 0.16 |
UNC5C |
unc-5 homolog C (C. elegans) |
133384 |
0.05 |
| chr20_56283949_56284203 | 0.16 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
740 |
0.76 |
| chr15_41202299_41202450 | 0.16 |
VPS18 |
vacuolar protein sorting 18 homolog (S. cerevisiae) |
15709 |
0.1 |
| chr10_50285208_50285359 | 0.15 |
VSTM4 |
V-set and transmembrane domain containing 4 |
38271 |
0.12 |
| chr20_35643588_35643790 | 0.15 |
RBL1 |
retinoblastoma-like 1 (p107) |
52769 |
0.13 |
| chr6_127839316_127839467 | 0.15 |
SOGA3 |
SOGA family member 3 |
645 |
0.78 |
| chr5_135365151_135365507 | 0.15 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
638 |
0.75 |
| chr5_39204467_39204671 | 0.15 |
FYB |
FYN binding protein |
1440 |
0.55 |
| chrX_103381032_103381192 | 0.15 |
SLC25A53 |
solute carrier family 25, member 53 |
20596 |
0.16 |
| chr11_76514494_76514645 | 0.15 |
RP11-21L23.2 |
|
3161 |
0.19 |
| chr11_119293301_119293628 | 0.15 |
THY1 |
Thy-1 cell surface antigen |
192 |
0.91 |
| chr3_65905504_65905655 | 0.15 |
ENSG00000264716 |
. |
21343 |
0.18 |
| chr7_94026391_94026542 | 0.15 |
COL1A2 |
collagen, type I, alpha 2 |
2593 |
0.39 |
| chr8_10000960_10001195 | 0.15 |
MSRA |
methionine sulfoxide reductase A |
47840 |
0.15 |
| chr2_109790043_109790205 | 0.15 |
ENSG00000264934 |
. |
32080 |
0.2 |
| chr13_20413212_20413363 | 0.14 |
RP11-61K9.2 |
|
2680 |
0.24 |
| chr22_37272954_37273105 | 0.14 |
NCF4 |
neutrophil cytosolic factor 4, 40kDa |
4850 |
0.15 |
| chr8_18694624_18694775 | 0.14 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
17185 |
0.22 |
| chr2_213279556_213279778 | 0.14 |
ENSG00000221782 |
. |
11417 |
0.25 |
| chr12_133770634_133770785 | 0.14 |
ZNF268 |
zinc finger protein 268 |
2626 |
0.19 |
| chr5_148793966_148794117 | 0.14 |
ENSG00000208035 |
. |
14440 |
0.12 |
| chr1_236147536_236147733 | 0.14 |
ENSG00000206803 |
. |
68919 |
0.09 |
| chr12_120241388_120241613 | 0.14 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
313 |
0.92 |
| chr19_3606102_3607092 | 0.14 |
TBXA2R |
thromboxane A2 receptor |
61 |
0.89 |
| chr9_115811689_115811840 | 0.14 |
ZFP37 |
ZFP37 zinc finger protein |
7204 |
0.26 |
| chr16_66223356_66223587 | 0.14 |
ENSG00000201999 |
. |
112241 |
0.06 |
| chr9_133655839_133655990 | 0.14 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
54539 |
0.13 |
| chr11_132863892_132864419 | 0.14 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
50492 |
0.18 |
| chr11_112125884_112126163 | 0.14 |
AP002884.2 |
LOC100132686 protein; Uncharacterized protein |
4965 |
0.13 |
| chr19_1361544_1361801 | 0.14 |
MUM1 |
melanoma associated antigen (mutated) 1 |
5309 |
0.1 |
| chr5_39211790_39211941 | 0.14 |
FYB |
FYN binding protein |
7800 |
0.29 |
| chr17_40933524_40933675 | 0.14 |
WNK4 |
WNK lysine deficient protein kinase 4 |
903 |
0.35 |
| chr12_12961145_12961613 | 0.14 |
DDX47 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 |
4871 |
0.18 |
| chr3_28958326_28958575 | 0.14 |
ENSG00000238470 |
. |
94283 |
0.09 |
| chr16_31022230_31022574 | 0.14 |
STX1B |
syntaxin 1B |
453 |
0.64 |
| chr6_56707059_56707245 | 0.14 |
DST |
dystonin |
791 |
0.6 |
| chr4_187622974_187623192 | 0.14 |
FAT1 |
FAT atypical cadherin 1 |
21926 |
0.25 |
| chrX_109246139_109246998 | 0.14 |
TMEM164 |
transmembrane protein 164 |
225 |
0.95 |
| chr2_183289157_183289308 | 0.14 |
PDE1A |
phosphodiesterase 1A, calmodulin-dependent |
2517 |
0.42 |
| chr19_41195400_41196566 | 0.14 |
NUMBL |
numb homolog (Drosophila)-like |
44 |
0.96 |
| chr18_34914843_34915294 | 0.14 |
RP11-797E24.3 |
|
60645 |
0.13 |
| chr12_111843881_111845308 | 0.14 |
SH2B3 |
SH2B adaptor protein 3 |
842 |
0.62 |
| chr1_178340504_178341006 | 0.13 |
RASAL2 |
RAS protein activator like 2 |
30149 |
0.22 |
| chr8_60000977_60001151 | 0.13 |
RP11-25K19.1 |
|
30535 |
0.17 |
| chrX_39964334_39965624 | 0.13 |
BCOR |
BCL6 corepressor |
8323 |
0.32 |
| chr7_90330496_90330666 | 0.13 |
CDK14 |
cyclin-dependent kinase 14 |
7999 |
0.29 |
| chrX_139608216_139608367 | 0.13 |
SOX3 |
SRY (sex determining region Y)-box 3 |
21066 |
0.26 |
| chr4_147576450_147576671 | 0.13 |
ENSG00000264323 |
. |
16147 |
0.21 |
| chr13_27135563_27135714 | 0.13 |
WASF3 |
WAS protein family, member 3 |
3751 |
0.35 |
| chr14_22938246_22938397 | 0.13 |
TRAJ60 |
T cell receptor alpha joining 60 (pseudogene) |
6975 |
0.11 |
| chrX_53349448_53350467 | 0.13 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
565 |
0.73 |
| chr11_64512499_64513555 | 0.13 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
99 |
0.95 |
| chr1_240629057_240629208 | 0.13 |
FMN2 |
formin 2 |
8350 |
0.21 |
| chr19_8675123_8675468 | 0.13 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
290 |
0.88 |
| chr11_69851843_69851994 | 0.13 |
ANO1-AS2 |
ANO1 antisense RNA 2 (head to head) |
69536 |
0.09 |
| chr16_23706248_23706462 | 0.13 |
ERN2 |
endoplasmic reticulum to nucleus signaling 2 |
274 |
0.86 |
| chr5_53810721_53810989 | 0.13 |
SNX18 |
sorting nexin 18 |
2734 |
0.38 |
| chr6_159235122_159235362 | 0.13 |
EZR-AS1 |
EZR antisense RNA 1 |
3801 |
0.21 |
| chr8_128957071_128957260 | 0.12 |
TMEM75 |
transmembrane protein 75 |
3426 |
0.25 |
| chr15_68872500_68872651 | 0.12 |
CORO2B |
coronin, actin binding protein, 2B |
1002 |
0.69 |
| chr7_152219742_152220164 | 0.12 |
ENSG00000199404 |
. |
70223 |
0.09 |
| chr12_88917218_88917369 | 0.12 |
KITLG |
KIT ligand |
56945 |
0.14 |
| chr7_100032883_100033657 | 0.12 |
RP11-758P17.2 |
|
390 |
0.45 |
| chr11_114175854_114176005 | 0.12 |
NNMT |
nicotinamide N-methyltransferase |
7056 |
0.22 |
| chr11_36052910_36053061 | 0.12 |
ENSG00000263389 |
. |
21337 |
0.21 |
| chr7_55545563_55545779 | 0.12 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
38099 |
0.19 |
| chr20_13229977_13230206 | 0.12 |
RP5-1077I2.3 |
|
4075 |
0.24 |
| chr14_77370360_77370542 | 0.12 |
ENSG00000223174 |
. |
13845 |
0.2 |
| chr19_3094656_3095580 | 0.12 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
710 |
0.53 |
| chr17_39969274_39969651 | 0.12 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
279 |
0.68 |
| chr20_33871790_33873092 | 0.12 |
EIF6 |
eukaryotic translation initiation factor 6 |
77 |
0.88 |
| chr12_104973868_104974019 | 0.12 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
8683 |
0.24 |
| chr1_201915320_201915636 | 0.12 |
LMOD1 |
leiomodin 1 (smooth muscle) |
237 |
0.87 |
| chr3_70881422_70881573 | 0.12 |
ENSG00000206939 |
. |
23996 |
0.24 |
| chr8_6693007_6693158 | 0.12 |
DEFB1 |
defensin, beta 1 |
42462 |
0.14 |
| chr8_101117971_101118266 | 0.12 |
RGS22 |
regulator of G-protein signaling 22 |
72 |
0.97 |
| chr12_12016436_12016587 | 0.12 |
ETV6 |
ets variant 6 |
22360 |
0.26 |
| chr17_29036450_29037561 | 0.12 |
ENSG00000241631 |
. |
7212 |
0.17 |
| chr8_93066568_93066719 | 0.12 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
8548 |
0.31 |
| chr8_77585853_77586055 | 0.12 |
ZFHX4 |
zinc finger homeobox 4 |
7500 |
0.24 |
| chr6_20680738_20680889 | 0.12 |
RP3-348I23.2 |
|
120112 |
0.06 |
| chr16_73248437_73248618 | 0.12 |
C16orf47 |
chromosome 16 open reading frame 47 |
70181 |
0.13 |
| chr19_45754620_45755932 | 0.12 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
726 |
0.57 |
| chr9_18476160_18476529 | 0.12 |
ADAMTSL1 |
ADAMTS-like 1 |
2113 |
0.45 |
| chr8_119390030_119390181 | 0.12 |
AC023590.1 |
Uncharacterized protein |
95624 |
0.09 |
| chr21_39720894_39721098 | 0.12 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
52148 |
0.17 |
| chr18_53537809_53537960 | 0.12 |
TCF4 |
transcription factor 4 |
205866 |
0.03 |
| chr15_49082630_49082781 | 0.11 |
RP11-485O10.2 |
|
7318 |
0.18 |
| chr1_101186979_101187209 | 0.11 |
VCAM1 |
vascular cell adhesion molecule 1 |
1774 |
0.47 |
| chr6_46458824_46459774 | 0.11 |
RCAN2 |
regulator of calcineurin 2 |
200 |
0.78 |
| chr12_57527559_57527864 | 0.11 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
1789 |
0.2 |
| chr6_106611529_106611909 | 0.11 |
RP1-134E15.3 |
|
63704 |
0.11 |
| chr19_14315927_14317134 | 0.11 |
LPHN1 |
latrophilin 1 |
451 |
0.77 |
| chr10_90711644_90712353 | 0.11 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
532 |
0.73 |
| chr15_101498489_101498640 | 0.11 |
LRRK1 |
leucine-rich repeat kinase 1 |
39014 |
0.15 |
| chr17_59361891_59362042 | 0.11 |
RP11-332H18.3 |
|
98180 |
0.06 |
| chr5_154033985_154034136 | 0.11 |
ENSG00000221552 |
. |
31276 |
0.14 |
| chr9_99277013_99277426 | 0.11 |
CDC14B |
cell division cycle 14B |
51893 |
0.13 |
| chr9_132033008_132033468 | 0.11 |
ENSG00000220992 |
. |
40242 |
0.13 |
| chr14_23000842_23001170 | 0.11 |
TRAJ15 |
T cell receptor alpha joining 15 |
2426 |
0.17 |
| chr1_25636296_25636447 | 0.11 |
TMEM50A |
transmembrane protein 50A |
28037 |
0.11 |
| chr11_3070813_3071033 | 0.11 |
ENSG00000201616 |
. |
1172 |
0.38 |
| chr17_42637473_42637723 | 0.11 |
FZD2 |
frizzled family receptor 2 |
2673 |
0.27 |
| chr19_47732183_47732562 | 0.11 |
BBC3 |
BCL2 binding component 3 |
2079 |
0.22 |
| chr6_158653547_158654353 | 0.11 |
ENSG00000265803 |
. |
57934 |
0.11 |
| chr2_121324572_121324723 | 0.11 |
ENSG00000201006 |
. |
84202 |
0.1 |
| chr5_95768778_95768929 | 0.11 |
PCSK1 |
proprotein convertase subtilisin/kexin type 1 |
131 |
0.98 |
| chr22_20105332_20105991 | 0.11 |
RANBP1 |
RAN binding protein 1 |
185 |
0.8 |
| chr9_101137213_101137445 | 0.11 |
TBC1D2 |
TBC1 domain family, member 2 |
119414 |
0.06 |
| chr10_123276154_123276305 | 0.11 |
FGFR2 |
fibroblast growth factor receptor 2 |
1536 |
0.55 |
| chr13_24826013_24826241 | 0.11 |
SPATA13 |
spermatogenesis associated 13 |
271 |
0.9 |
| chr12_92270812_92270963 | 0.11 |
C12orf79 |
chromosome 12 open reading frame 79 |
259910 |
0.02 |
| chr12_124427345_124427647 | 0.11 |
CCDC92 |
coiled-coil domain containing 92 |
3824 |
0.09 |
| chr18_22868744_22868981 | 0.11 |
CTD-2006O16.2 |
|
13228 |
0.28 |
| chr12_31783207_31783359 | 0.11 |
METTL20 |
methyltransferase like 20 |
16811 |
0.15 |
| chr6_3340953_3341317 | 0.11 |
RP11-506K6.4 |
|
29239 |
0.19 |
| chr12_11916647_11916798 | 0.11 |
ETV6 |
ets variant 6 |
11287 |
0.29 |
| chr11_76382331_76382482 | 0.11 |
LRRC32 |
leucine rich repeat containing 32 |
615 |
0.71 |
| chr16_27256695_27256846 | 0.11 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
12371 |
0.16 |
| chr5_134178464_134178621 | 0.11 |
C5orf24 |
chromosome 5 open reading frame 24 |
2828 |
0.25 |
| chr12_93789120_93789411 | 0.11 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
16835 |
0.14 |
| chr11_19735768_19735975 | 0.11 |
NAV2 |
neuron navigator 2 |
728 |
0.74 |
| chr2_171362653_171362804 | 0.11 |
AC007277.3 |
|
137726 |
0.05 |
| chr6_143164322_143164558 | 0.11 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
6256 |
0.3 |
| chr1_161167282_161167469 | 0.11 |
ADAMTS4 |
ADAM metallopeptidase with thrombospondin type 1 motif, 4 |
1468 |
0.16 |
| chr11_114168512_114168913 | 0.11 |
NNMT |
nicotinamide N-methyltransferase |
106 |
0.97 |
| chr5_143407637_143407837 | 0.11 |
ENSG00000239390 |
. |
112707 |
0.07 |
| chr18_18549206_18549357 | 0.11 |
ROCK1 |
Rho-associated, coiled-coil containing protein kinase 1 |
14450 |
0.28 |
| chr13_60709592_60709946 | 0.11 |
DIAPH3-AS2 |
DIAPH3 antisense RNA 2 |
9063 |
0.22 |
| chr11_123300349_123300500 | 0.10 |
AP000783.1 |
Uncharacterized protein |
634 |
0.77 |
| chr11_1893200_1893351 | 0.10 |
LSP1 |
lymphocyte-specific protein 1 |
1159 |
0.31 |
| chr1_29308973_29309136 | 0.10 |
ENSG00000206704 |
. |
3168 |
0.29 |
| chr11_2402201_2402352 | 0.10 |
CD81 |
CD81 molecule |
1414 |
0.26 |
| chr15_48927305_48927456 | 0.10 |
FBN1 |
fibrillin 1 |
10538 |
0.28 |
| chr22_17671940_17672186 | 0.10 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
8610 |
0.17 |
| chr5_34622373_34622524 | 0.10 |
RAI14 |
retinoic acid induced 14 |
33894 |
0.19 |
| chr21_36936389_36936667 | 0.10 |
ENSG00000211590 |
. |
156485 |
0.04 |
| chr1_113087859_113088010 | 0.10 |
RP4-671G15.2 |
|
26871 |
0.14 |
| chr19_46148736_46148888 | 0.10 |
EML2 |
echinoderm microtubule associated protein like 2 |
75 |
0.93 |
| chr22_25346545_25346696 | 0.10 |
KIAA1671 |
KIAA1671 |
2077 |
0.32 |
| chr13_52158653_52159782 | 0.10 |
WDFY2 |
WD repeat and FYVE domain containing 2 |
573 |
0.6 |
| chr2_109202767_109202918 | 0.10 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
1925 |
0.4 |
| chrX_54555990_54556466 | 0.10 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
416 |
0.88 |
| chr16_87978942_87979129 | 0.10 |
BANP |
BTG3 associated nuclear protein |
5007 |
0.21 |
| chr4_169616116_169616373 | 0.10 |
ENSG00000206613 |
. |
8377 |
0.21 |
| chr1_156215307_156216024 | 0.10 |
PAQR6 |
progestin and adipoQ receptor family member VI |
2157 |
0.16 |
| chr14_101867418_101867569 | 0.10 |
ENSG00000258498 |
. |
159266 |
0.02 |
| chr13_33850317_33850468 | 0.10 |
ENSG00000236581 |
. |
1298 |
0.49 |
| chrY_7395772_7396011 | 0.10 |
ENSG00000252472 |
. |
104692 |
0.07 |
| chr16_72955812_72956039 | 0.10 |
ENSG00000221799 |
. |
62831 |
0.11 |
| chr13_36492055_36492221 | 0.10 |
DCLK1 |
doublecortin-like kinase 1 |
62359 |
0.15 |
| chr10_25971446_25971646 | 0.10 |
ENSG00000223019 |
. |
131300 |
0.05 |
| chr14_99699001_99699256 | 0.10 |
AL109767.1 |
|
30157 |
0.18 |
| chr19_8656644_8657344 | 0.10 |
ADAMTS10 |
ADAM metallopeptidase with thrombospondin type 1 motif, 10 |
86 |
0.96 |
| chr2_231090060_231090211 | 0.10 |
SP110 |
SP110 nuclear body protein |
309 |
0.48 |
| chr2_219646650_219647418 | 0.10 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
555 |
0.7 |
| chr10_36739274_36739523 | 0.10 |
ENSG00000237002 |
. |
11809 |
0.29 |
| chr20_32030739_32031661 | 0.10 |
SNTA1 |
syntrophin, alpha 1 |
498 |
0.77 |
| chr22_30641977_30642328 | 0.10 |
RP1-102K2.8 |
|
42 |
0.9 |
| chr1_36042987_36043882 | 0.10 |
RP4-728D4.2 |
|
104 |
0.95 |
| chr15_60688893_60689153 | 0.10 |
ANXA2 |
annexin A2 |
514 |
0.85 |
| chr1_78005375_78005526 | 0.10 |
AK5 |
adenylate kinase 5 |
7658 |
0.25 |
| chr15_65197450_65197821 | 0.10 |
ENSG00000264929 |
. |
5197 |
0.16 |
| chr19_17445022_17445529 | 0.10 |
ANO8 |
anoctamin 8 |
363 |
0.48 |
| chr6_43214536_43215130 | 0.10 |
TTBK1 |
tau tubulin kinase 1 |
894 |
0.48 |
| chr10_124254663_124255115 | 0.10 |
HTRA1 |
HtrA serine peptidase 1 |
11318 |
0.2 |
| chr5_121416588_121416739 | 0.10 |
LOX |
lysyl oxidase |
2683 |
0.32 |
| chr6_36593181_36593356 | 0.10 |
ENSG00000263894 |
. |
2979 |
0.19 |
| chr14_52346903_52347054 | 0.10 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
2668 |
0.28 |
| chr12_77309523_77309699 | 0.10 |
CSRP2 |
cysteine and glycine-rich protein 2 |
36771 |
0.18 |
| chr2_3497402_3497553 | 0.10 |
ADI1 |
acireductone dioxygenase 1 |
7110 |
0.15 |
| chr10_88162500_88162651 | 0.10 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
36340 |
0.16 |
| chr1_211589992_211590552 | 0.10 |
TRAF5 |
TNF receptor-associated factor 5 |
70566 |
0.1 |
| chr9_118919385_118919577 | 0.10 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
3398 |
0.32 |
| chr7_27404330_27404615 | 0.10 |
EVX1-AS |
EVX1 antisense RNA |
117624 |
0.04 |
| chr22_41956802_41957044 | 0.10 |
CSDC2 |
cold shock domain containing C2, RNA binding |
156 |
0.93 |
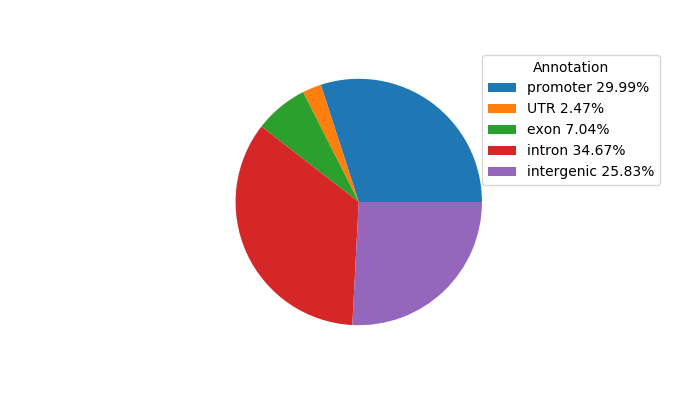
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.0 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0071883 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:1903077 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.0 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.0 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0009415 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.0 | GO:2001012 | mesenchymal cell differentiation involved in kidney development(GO:0072161) metanephric mesenchymal cell differentiation(GO:0072162) mesenchymal cell differentiation involved in renal system development(GO:2001012) |
| 0.0 | 0.1 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.0 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.0 | 0.1 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.0 | GO:1901889 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.0 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |