Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
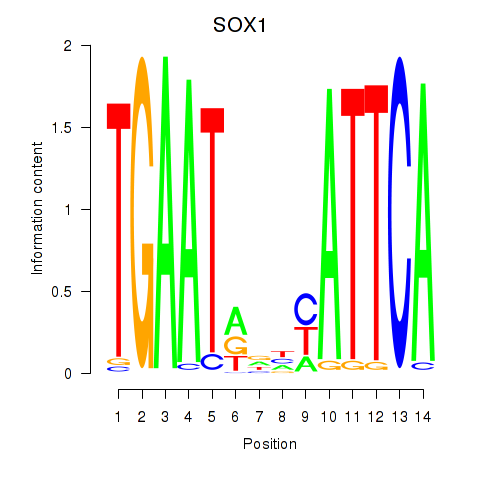
Results for SOX1
Z-value: 0.98
Transcription factors associated with SOX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX1
|
ENSG00000182968.3 | SRY-box transcription factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_112722136_112722317 | SOX1 | 313 | 0.924270 | 0.44 | 2.3e-01 | Click! |
| chr13_112720584_112720950 | SOX1 | 1146 | 0.580299 | -0.40 | 2.8e-01 | Click! |
| chr13_112797044_112797195 | SOX1 | 75206 | 0.112819 | -0.24 | 5.4e-01 | Click! |
| chr13_112721079_112721230 | SOX1 | 759 | 0.729661 | 0.10 | 8.0e-01 | Click! |
| chr13_112770858_112771009 | SOX1 | 49020 | 0.166793 | 0.05 | 9.0e-01 | Click! |
Activity of the SOX1 motif across conditions
Conditions sorted by the z-value of the SOX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
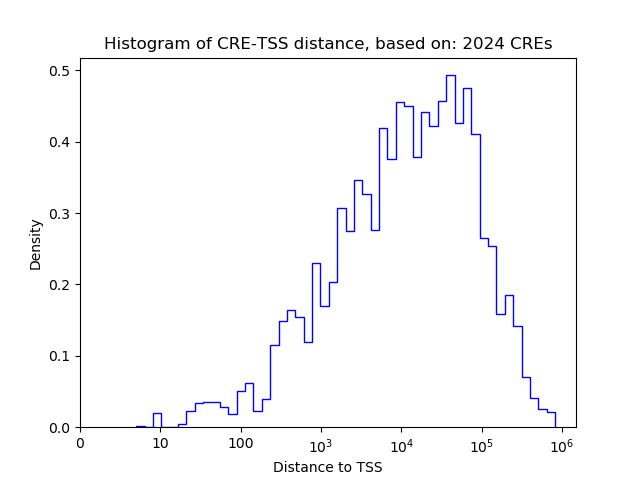
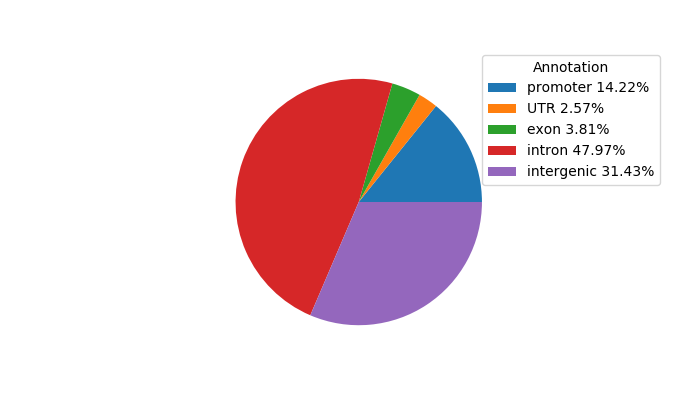
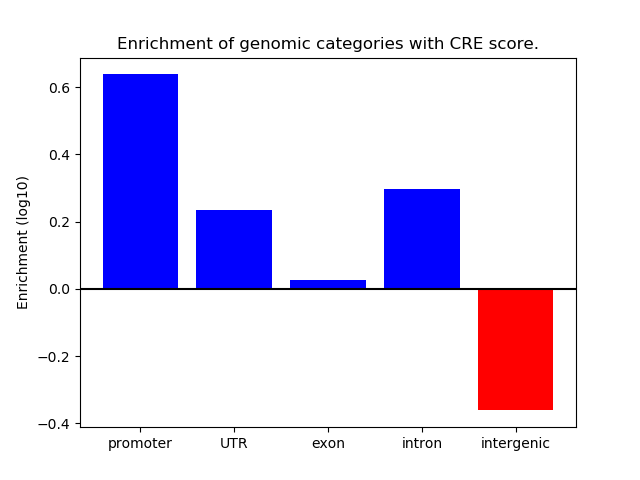
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_1772879_1773230 | 0.67 |
GNB1 |
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
9330 |
0.15 |
| chr13_101046001_101046242 | 0.42 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
25368 |
0.23 |
| chr10_60273912_60274168 | 0.42 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
1140 |
0.65 |
| chr1_186072218_186072510 | 0.41 |
HMCN1 |
hemicentin 1 |
68155 |
0.12 |
| chr14_27065632_27065895 | 0.41 |
NOVA1 |
neuro-oncological ventral antigen 1 |
482 |
0.74 |
| chr4_75718841_75720131 | 0.40 |
BTC |
betacellulin |
410 |
0.91 |
| chr1_157964729_157965124 | 0.39 |
KIRREL |
kin of IRRE like (Drosophila) |
1491 |
0.45 |
| chr17_56744273_56744616 | 0.36 |
ENSG00000199426 |
. |
386 |
0.76 |
| chr2_33360506_33360697 | 0.34 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
877 |
0.73 |
| chr11_46300696_46300847 | 0.33 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
1543 |
0.37 |
| chr6_85823864_85824548 | 0.33 |
NT5E |
5'-nucleotidase, ecto (CD73) |
335603 |
0.01 |
| chr4_127699042_127699253 | 0.33 |
ENSG00000199862 |
. |
250227 |
0.02 |
| chr4_134068181_134068851 | 0.33 |
PCDH10 |
protocadherin 10 |
1954 |
0.52 |
| chr5_24644888_24645378 | 0.32 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
46 |
0.99 |
| chr22_29372984_29373407 | 0.32 |
ZNRF3 |
zinc and ring finger 3 |
9845 |
0.17 |
| chr11_71780711_71781020 | 0.31 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
268 |
0.8 |
| chr9_40632604_40633916 | 0.30 |
SPATA31A3 |
SPATA31 subfamily A, member 3 |
67031 |
0.14 |
| chr8_107985745_107985896 | 0.30 |
ABRA |
actin-binding Rho activating protein |
203347 |
0.03 |
| chr4_113810751_113811050 | 0.30 |
RP11-119H12.6 |
|
7414 |
0.26 |
| chr15_30112674_30113382 | 0.29 |
TJP1 |
tight junction protein 1 |
723 |
0.64 |
| chr1_61332602_61332753 | 0.29 |
NFIA |
nuclear factor I/A |
1746 |
0.51 |
| chr2_33802935_33803086 | 0.29 |
AC020594.5 |
|
14396 |
0.22 |
| chr15_92404681_92404832 | 0.29 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
7405 |
0.27 |
| chr2_227658957_227659437 | 0.29 |
IRS1 |
insulin receptor substrate 1 |
5278 |
0.23 |
| chr8_104121485_104121788 | 0.28 |
KB-1639H6.4 |
|
11624 |
0.13 |
| chr5_32027565_32028229 | 0.28 |
ENSG00000266243 |
. |
91632 |
0.07 |
| chr1_162603783_162603934 | 0.28 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
1598 |
0.41 |
| chr9_65576390_65577630 | 0.27 |
SPATA31A7 |
SPATA31 subfamily A, member 7 |
67400 |
0.15 |
| chr5_127398296_127398447 | 0.27 |
SLC12A2 |
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
21087 |
0.29 |
| chr3_112354153_112354320 | 0.27 |
CCDC80 |
coiled-coil domain containing 80 |
2708 |
0.34 |
| chr2_55338658_55339041 | 0.27 |
RTN4 |
reticulon 4 |
908 |
0.57 |
| chr17_74442541_74443075 | 0.27 |
UBE2O |
ubiquitin-conjugating enzyme E2O |
6480 |
0.12 |
| chr3_104078695_104079148 | 0.27 |
ENSG00000265076 |
. |
305281 |
0.01 |
| chr4_5749568_5749826 | 0.27 |
EVC |
Ellis van Creveld syndrome |
36755 |
0.18 |
| chr14_89020611_89020920 | 0.26 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
312 |
0.86 |
| chrX_139792263_139792666 | 0.26 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
532 |
0.82 |
| chr13_25883872_25884023 | 0.26 |
NUPL1 |
nucleoporin like 1 |
1987 |
0.33 |
| chr14_31022770_31022975 | 0.26 |
G2E3 |
G2/M-phase specific E3 ubiquitin protein ligase |
5457 |
0.25 |
| chr5_146887879_146888030 | 0.26 |
DPYSL3 |
dihydropyrimidinase-like 3 |
1665 |
0.51 |
| chr6_108971810_108972003 | 0.26 |
FOXO3 |
forkhead box O3 |
5643 |
0.32 |
| chr15_101783743_101783976 | 0.26 |
CHSY1 |
chondroitin sulfate synthase 1 |
8278 |
0.18 |
| chr6_102219183_102219334 | 0.26 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
46987 |
0.21 |
| chr2_229517064_229517215 | 0.26 |
ENSG00000251801 |
. |
64550 |
0.15 |
| chr7_94029901_94030156 | 0.25 |
COL1A2 |
collagen, type I, alpha 2 |
6155 |
0.29 |
| chr3_61623550_61623701 | 0.25 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
76040 |
0.12 |
| chr12_72671108_72671475 | 0.25 |
ENSG00000236333 |
. |
2604 |
0.33 |
| chr18_55671489_55671692 | 0.25 |
ENSG00000265200 |
. |
12526 |
0.24 |
| chr1_240760211_240760638 | 0.24 |
RP11-467I20.6 |
|
6376 |
0.21 |
| chr21_17568279_17568430 | 0.24 |
ENSG00000201025 |
. |
88735 |
0.1 |
| chr4_148693225_148693748 | 0.24 |
ENSG00000264274 |
. |
10260 |
0.18 |
| chr1_170556453_170556642 | 0.24 |
RP11-576I22.2 |
|
54759 |
0.13 |
| chr15_99366541_99366815 | 0.24 |
ENSG00000264480 |
. |
39023 |
0.17 |
| chr5_140863842_140863993 | 0.24 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
824 |
0.39 |
| chr1_153584096_153584410 | 0.23 |
S100A16 |
S100 calcium binding protein A16 |
1306 |
0.22 |
| chr9_100181817_100181968 | 0.23 |
TDRD7 |
tudor domain containing 7 |
7516 |
0.23 |
| chr5_176783043_176783194 | 0.23 |
RGS14 |
regulator of G-protein signaling 14 |
1720 |
0.2 |
| chr2_216297601_216297993 | 0.23 |
FN1 |
fibronectin 1 |
2993 |
0.27 |
| chrX_53080432_53080669 | 0.23 |
GPR173 |
G protein-coupled receptor 173 |
2085 |
0.31 |
| chr4_170063887_170064038 | 0.23 |
RP11-327O17.2 |
|
58983 |
0.13 |
| chr7_18794470_18794725 | 0.23 |
ENSG00000222164 |
. |
53305 |
0.17 |
| chr3_177333090_177333241 | 0.23 |
ENSG00000200288 |
. |
9032 |
0.31 |
| chr15_75335743_75336025 | 0.23 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
268 |
0.9 |
| chr18_53089837_53090372 | 0.23 |
TCF4 |
transcription factor 4 |
361 |
0.89 |
| chr4_153272766_153273041 | 0.23 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
1220 |
0.52 |
| chr20_43645221_43645372 | 0.23 |
ENSG00000252021 |
. |
12307 |
0.16 |
| chr20_19959113_19959531 | 0.22 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
38438 |
0.16 |
| chr20_39944817_39945241 | 0.22 |
ZHX3 |
zinc fingers and homeoboxes 3 |
797 |
0.64 |
| chr15_57350079_57350487 | 0.22 |
ENSG00000222586 |
. |
91688 |
0.09 |
| chr19_56118074_56118225 | 0.22 |
ZNF865 |
zinc finger protein 865 |
1378 |
0.19 |
| chr6_86113632_86113933 | 0.22 |
NT5E |
5'-nucleotidase, ecto (CD73) |
46027 |
0.19 |
| chr17_66706567_66706718 | 0.22 |
ENSG00000263690 |
. |
56058 |
0.14 |
| chr12_42982187_42982354 | 0.22 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
1208 |
0.6 |
| chr6_138867193_138867344 | 0.22 |
NHSL1 |
NHS-like 1 |
420 |
0.9 |
| chr6_27261362_27261571 | 0.22 |
POM121L2 |
POM121 transmembrane nucleoporin-like 2 |
17625 |
0.19 |
| chr8_116519428_116519579 | 0.22 |
AF178030.2 |
|
2326 |
0.38 |
| chr21_44496501_44497065 | 0.22 |
CBS |
cystathionine-beta-synthase |
270 |
0.91 |
| chr4_77508066_77508217 | 0.22 |
ENSG00000265314 |
. |
11437 |
0.17 |
| chr2_145189034_145189185 | 0.22 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
972 |
0.69 |
| chr5_96433895_96434046 | 0.22 |
CTD-2215E18.1 |
Uncharacterized protein |
9191 |
0.21 |
| chr1_192780744_192780895 | 0.22 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
2648 |
0.37 |
| chr2_36587826_36587983 | 0.22 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
4290 |
0.35 |
| chr1_119627265_119627416 | 0.22 |
WARS2-IT1 |
WARS2 intronic transcript 1 (non-protein coding) |
37312 |
0.16 |
| chrX_139844529_139844680 | 0.22 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
22119 |
0.21 |
| chr3_525379_525530 | 0.22 |
CHL1 |
cell adhesion molecule L1-like |
94326 |
0.09 |
| chr20_61656853_61657255 | 0.22 |
BHLHE23 |
basic helix-loop-helix family, member e23 |
18667 |
0.17 |
| chr7_36301220_36301371 | 0.21 |
AC007327.5 |
|
30579 |
0.15 |
| chr14_42071339_42071490 | 0.21 |
LRFN5 |
leucine rich repeat and fibronectin type III domain containing 5 |
5359 |
0.27 |
| chr7_93465582_93465846 | 0.21 |
TFPI2 |
tissue factor pathway inhibitor 2 |
53768 |
0.12 |
| chr18_53537196_53537347 | 0.21 |
TCF4 |
transcription factor 4 |
205253 |
0.03 |
| chr1_26436861_26437012 | 0.21 |
PDIK1L |
PDLIM1 interacting kinase 1 like |
728 |
0.54 |
| chr6_56555605_56555856 | 0.21 |
DST |
dystonin |
47936 |
0.18 |
| chr5_100163193_100163344 | 0.21 |
ENSG00000221263 |
. |
10999 |
0.28 |
| chr7_18125753_18126403 | 0.21 |
HDAC9 |
histone deacetylase 9 |
494 |
0.84 |
| chr1_200982094_200982245 | 0.21 |
KIF21B |
kinesin family member 21B |
10367 |
0.19 |
| chr19_44661345_44661496 | 0.21 |
ZNF226 |
zinc finger protein 226 |
7806 |
0.12 |
| chr8_116463339_116463609 | 0.21 |
TRPS1 |
trichorhinophalangeal syndrome I |
40974 |
0.2 |
| chr1_36834692_36834843 | 0.21 |
STK40 |
serine/threonine kinase 40 |
7826 |
0.13 |
| chr13_36045598_36045825 | 0.21 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
5121 |
0.22 |
| chr8_24931073_24931224 | 0.21 |
ENSG00000241811 |
. |
108087 |
0.06 |
| chr20_25506914_25507184 | 0.21 |
NINL |
ninein-like |
47386 |
0.13 |
| chr13_80912572_80913077 | 0.21 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
970 |
0.67 |
| chr2_200168941_200169130 | 0.21 |
RP11-486F17.1 |
|
20998 |
0.26 |
| chr1_114862373_114862591 | 0.21 |
TRIM33 |
tripartite motif containing 33 |
143376 |
0.04 |
| chr5_122452023_122452174 | 0.21 |
AC106786.1 |
|
26104 |
0.15 |
| chr20_17539236_17539459 | 0.21 |
BFSP1 |
beaded filament structural protein 1, filensin |
136 |
0.96 |
| chr9_109624062_109624226 | 0.21 |
ZNF462 |
zinc finger protein 462 |
1234 |
0.53 |
| chr2_29751932_29752128 | 0.21 |
ENSG00000266310 |
. |
25171 |
0.26 |
| chr8_97350778_97351015 | 0.20 |
ENSG00000199732 |
. |
29346 |
0.17 |
| chr15_92722178_92722335 | 0.20 |
RP11-24J19.1 |
|
6590 |
0.32 |
| chr2_237653364_237653686 | 0.20 |
ACKR3 |
atypical chemokine receptor 3 |
175241 |
0.03 |
| chr4_187648163_187648374 | 0.20 |
FAT1 |
FAT atypical cadherin 1 |
392 |
0.92 |
| chr2_70151329_70151480 | 0.20 |
MXD1 |
MAX dimerization protein 1 |
9084 |
0.12 |
| chr9_100010529_100010680 | 0.20 |
CCDC180 |
coiled-coil domain containing 180 |
9825 |
0.25 |
| chr6_97663203_97663434 | 0.20 |
MMS22L |
MMS22-like, DNA repair protein |
47910 |
0.19 |
| chr6_121843994_121844420 | 0.20 |
ENSG00000201379 |
. |
19565 |
0.2 |
| chr14_36560741_36560892 | 0.20 |
ENSG00000212071 |
. |
104958 |
0.07 |
| chr10_32229562_32229783 | 0.20 |
ARHGAP12 |
Rho GTPase activating protein 12 |
11930 |
0.24 |
| chr5_167079523_167079674 | 0.20 |
CTB-78F1.1 |
|
7888 |
0.28 |
| chr5_84589012_84589394 | 0.20 |
ENSG00000215953 |
. |
234749 |
0.02 |
| chr21_44499975_44500459 | 0.20 |
CBS |
cystathionine-beta-synthase |
3164 |
0.22 |
| chr14_75909783_75910034 | 0.20 |
JDP2 |
Jun dimerization protein 2 |
11071 |
0.19 |
| chr5_124536368_124536614 | 0.20 |
ENSG00000222107 |
. |
150333 |
0.05 |
| chr7_157918330_157918481 | 0.20 |
AC011899.10 |
|
98340 |
0.08 |
| chr10_17074110_17074261 | 0.20 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
47991 |
0.17 |
| chr3_114137011_114137162 | 0.20 |
ZBTB20-AS1 |
ZBTB20 antisense RNA 1 |
30570 |
0.18 |
| chr5_39523911_39524062 | 0.19 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
61584 |
0.15 |
| chr2_196978205_196978455 | 0.19 |
RP11-347P5.1 |
|
37657 |
0.15 |
| chr9_82093653_82093804 | 0.19 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
92960 |
0.1 |
| chr16_11930217_11930382 | 0.19 |
RSL1D1 |
ribosomal L1 domain containing 1 |
14603 |
0.12 |
| chr16_84927762_84927913 | 0.19 |
ENSG00000265249 |
. |
41263 |
0.14 |
| chrX_57933599_57933750 | 0.19 |
ZXDA |
zinc finger, X-linked, duplicated A |
3393 |
0.4 |
| chr17_2309940_2310157 | 0.19 |
MNT |
MAX network transcriptional repressor |
5636 |
0.12 |
| chr19_58055939_58056090 | 0.19 |
ZNF550 |
zinc finger protein 550 |
3347 |
0.13 |
| chr7_84121446_84121597 | 0.19 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
344 |
0.94 |
| chr4_139360491_139360642 | 0.19 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
197063 |
0.03 |
| chr12_120697381_120697532 | 0.19 |
PXN |
paxillin |
6107 |
0.13 |
| chr1_35754106_35754533 | 0.19 |
ENSG00000240374 |
. |
3780 |
0.18 |
| chr5_17743873_17744038 | 0.19 |
ENSG00000201715 |
. |
398230 |
0.01 |
| chr1_61668583_61669016 | 0.19 |
RP4-802A10.1 |
|
78394 |
0.11 |
| chr5_124042484_124042635 | 0.19 |
RP11-436H11.2 |
|
21965 |
0.13 |
| chr3_156220748_156220899 | 0.19 |
RP11-305K5.1 |
|
21213 |
0.21 |
| chr5_41790406_41790557 | 0.19 |
OXCT1 |
3-oxoacid CoA transferase 1 |
4001 |
0.32 |
| chr12_81258392_81258564 | 0.19 |
LIN7A |
lin-7 homolog A (C. elegans) |
24649 |
0.18 |
| chr8_42712037_42712188 | 0.19 |
THAP1 |
THAP domain containing, apoptosis associated protein 1 |
13644 |
0.14 |
| chr7_116169535_116169759 | 0.19 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
3300 |
0.2 |
| chrX_114851642_114851793 | 0.19 |
PLS3 |
plastin 3 |
7162 |
0.23 |
| chr4_47838472_47839750 | 0.19 |
CORIN |
corin, serine peptidase |
855 |
0.65 |
| chr19_1415408_1415673 | 0.19 |
CTB-25B13.13 |
|
1640 |
0.16 |
| chr2_200320961_200321297 | 0.19 |
SATB2 |
SATB homeobox 2 |
318 |
0.86 |
| chr9_43685064_43685767 | 0.19 |
CNTNAP3B |
contactin associated protein-like 3B |
505 |
0.86 |
| chr15_49916935_49917216 | 0.19 |
DTWD1 |
DTW domain containing 1 |
3779 |
0.23 |
| chr8_19148668_19149118 | 0.19 |
SH2D4A |
SH2 domain containing 4A |
22235 |
0.28 |
| chr3_106447792_106448185 | 0.19 |
ENSG00000200361 |
. |
40463 |
0.22 |
| chr6_24858289_24858440 | 0.19 |
ENSG00000263391 |
. |
18071 |
0.17 |
| chr1_202938451_202938746 | 0.19 |
CYB5R1 |
cytochrome b5 reductase 1 |
2190 |
0.2 |
| chr8_57086200_57086351 | 0.19 |
PLAG1 |
pleiomorphic adenoma gene 1 |
37563 |
0.12 |
| chr2_189839524_189839773 | 0.18 |
COL3A1 |
collagen, type III, alpha 1 |
549 |
0.79 |
| chr9_12859535_12860149 | 0.18 |
ENSG00000222658 |
. |
25478 |
0.2 |
| chr7_19308836_19308987 | 0.18 |
FERD3L |
Fer3-like bHLH transcription factor |
123867 |
0.05 |
| chr12_76144948_76145099 | 0.18 |
ENSG00000264062 |
. |
26702 |
0.22 |
| chr10_101952384_101952535 | 0.18 |
RP11-316M21.7 |
|
2666 |
0.19 |
| chr6_56507492_56507755 | 0.18 |
DST |
dystonin |
35 |
0.99 |
| chr15_33016455_33016606 | 0.18 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
6325 |
0.22 |
| chr15_96861717_96861868 | 0.18 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
4082 |
0.19 |
| chr1_200121796_200121947 | 0.18 |
ENSG00000221403 |
. |
7909 |
0.28 |
| chr7_115967399_115967904 | 0.18 |
AC073130.3 |
|
299 |
0.91 |
| chr1_65399507_65400011 | 0.18 |
JAK1 |
Janus kinase 1 |
32428 |
0.2 |
| chr2_8553463_8553614 | 0.18 |
AC011747.7 |
|
262358 |
0.02 |
| chr1_186970310_186970482 | 0.18 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
172274 |
0.04 |
| chr13_72377548_72377699 | 0.18 |
DACH1 |
dachshund homolog 1 (Drosophila) |
63284 |
0.15 |
| chr8_64131982_64132133 | 0.18 |
ENSG00000241964 |
. |
15412 |
0.23 |
| chr2_27010559_27010710 | 0.18 |
CENPA |
centromere protein A |
1698 |
0.35 |
| chr6_130554708_130554859 | 0.18 |
SAMD3 |
sterile alpha motif domain containing 3 |
10684 |
0.28 |
| chr5_124821676_124821827 | 0.18 |
ENSG00000222107 |
. |
134927 |
0.06 |
| chr12_76144582_76144847 | 0.18 |
ENSG00000264062 |
. |
26393 |
0.22 |
| chr1_81703498_81704139 | 0.18 |
ENSG00000223026 |
. |
13656 |
0.28 |
| chr10_34686084_34686235 | 0.18 |
PARD3 |
par-3 family cell polarity regulator |
24698 |
0.28 |
| chr6_46702576_46703631 | 0.18 |
PLA2G7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
24 |
0.97 |
| chr14_52120103_52120254 | 0.18 |
FRMD6 |
FERM domain containing 6 |
1480 |
0.42 |
| chr9_126978620_126978771 | 0.18 |
NEK6 |
NIMA-related kinase 6 |
41190 |
0.14 |
| chr11_2972331_2972482 | 0.18 |
ENSG00000207008 |
. |
12717 |
0.12 |
| chr2_66664825_66665240 | 0.18 |
MEIS1 |
Meis homeobox 1 |
93 |
0.96 |
| chr17_46634239_46634480 | 0.18 |
HOXB-AS2 |
HOXB cluster antisense RNA 2 |
265 |
0.61 |
| chr12_68915355_68915540 | 0.18 |
RAP1B |
RAP1B, member of RAS oncogene family |
89172 |
0.08 |
| chr5_159722948_159723245 | 0.18 |
ENSG00000243654 |
. |
4301 |
0.21 |
| chr11_13204755_13204972 | 0.18 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
93336 |
0.09 |
| chr17_75316367_75316959 | 0.18 |
SEPT9 |
septin 9 |
268 |
0.93 |
| chr8_121377938_121378124 | 0.18 |
COL14A1 |
collagen, type XIV, alpha 1 |
20346 |
0.24 |
| chr1_109934140_109934542 | 0.17 |
SORT1 |
sortilin 1 |
1638 |
0.33 |
| chr21_14607521_14607672 | 0.17 |
AL050302.1 |
Uncharacterized protein |
137790 |
0.05 |
| chr8_56806111_56806262 | 0.17 |
RP11-318K15.2 |
|
32 |
0.97 |
| chr17_53799692_53799908 | 0.17 |
TMEM100 |
transmembrane protein 100 |
417 |
0.89 |
| chr8_37378833_37379079 | 0.17 |
RP11-150O12.6 |
|
4417 |
0.31 |
| chr22_28741040_28741191 | 0.17 |
ENSG00000199797 |
. |
41373 |
0.19 |
| chr7_24988449_24988600 | 0.17 |
OSBPL3 |
oxysterol binding protein-like 3 |
30812 |
0.21 |
| chr15_35950174_35950564 | 0.17 |
DPH6 |
diphthamine biosynthesis 6 |
111976 |
0.07 |
| chr1_38317055_38317206 | 0.17 |
MTF1 |
metal-regulatory transcription factor 1 |
8162 |
0.1 |
| chr22_40578675_40578895 | 0.17 |
TNRC6B |
trinucleotide repeat containing 6B |
4839 |
0.3 |
| chr9_113598070_113598221 | 0.17 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
60518 |
0.13 |
| chr9_124329295_124329519 | 0.17 |
DAB2IP |
DAB2 interacting protein |
8 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.1 | 0.2 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.2 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.2 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.3 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.1 | GO:0043931 | ossification involved in bone maturation(GO:0043931) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.2 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0070091 | glucagon secretion(GO:0070091) |
| 0.0 | 0.2 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.0 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.0 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0030669 | clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |