Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
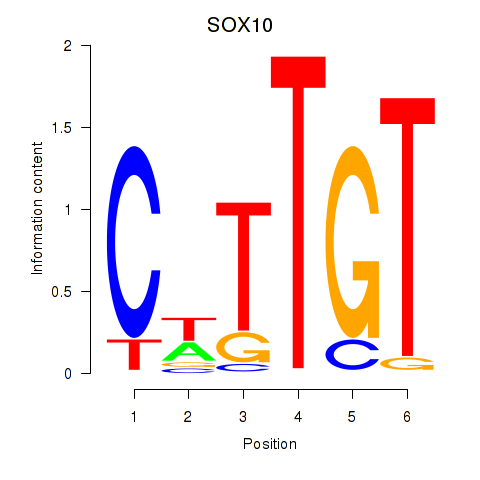
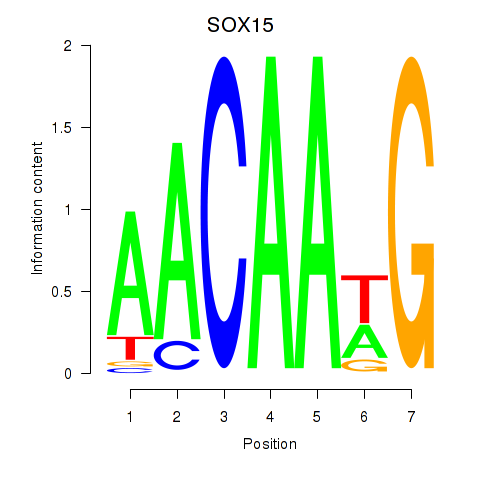
Results for SOX10_SOX15
Z-value: 0.40
Transcription factors associated with SOX10_SOX15
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX10
|
ENSG00000100146.12 | SRY-box transcription factor 10 |
|
SOX15
|
ENSG00000129194.3 | SRY-box transcription factor 15 |
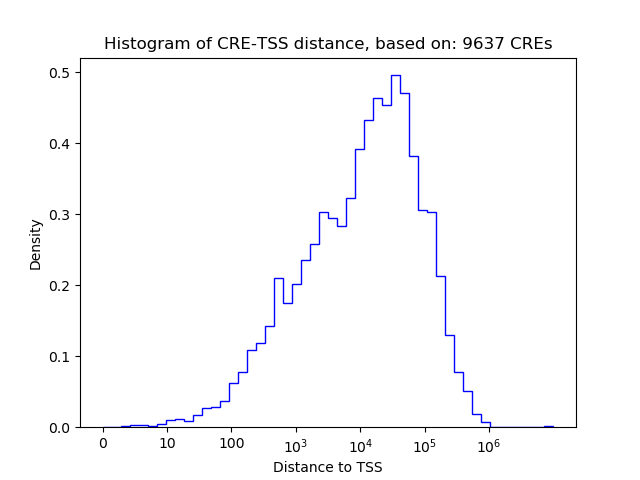
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_7494301_7494452 | SOX15 | 888 | 0.243344 | -0.56 | 1.2e-01 | Click! |
| chr17_7493173_7493324 | SOX15 | 142 | 0.838372 | -0.48 | 1.9e-01 | Click! |
| chr17_7492744_7493045 | SOX15 | 496 | 0.463197 | -0.41 | 2.8e-01 | Click! |
| chr17_7491214_7491503 | SOX15 | 2032 | 0.090549 | -0.29 | 4.5e-01 | Click! |
| chr17_7492081_7492232 | SOX15 | 1234 | 0.158874 | -0.21 | 5.8e-01 | Click! |
Activity of the SOX10_SOX15 motif across conditions
Conditions sorted by the z-value of the SOX10_SOX15 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
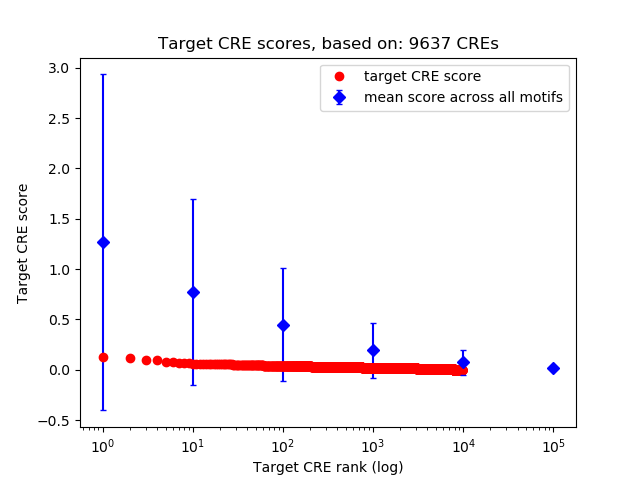
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_27218725_27218915 | 0.12 |
RP1-170O19.20 |
Uncharacterized protein |
812 |
0.27 |
| chr4_81195528_81195918 | 0.12 |
FGF5 |
fibroblast growth factor 5 |
7930 |
0.26 |
| chr14_73881950_73882101 | 0.10 |
NUMB |
numb homolog (Drosophila) |
5246 |
0.21 |
| chr9_18476160_18476529 | 0.09 |
ADAMTSL1 |
ADAMTS-like 1 |
2113 |
0.45 |
| chr12_91721216_91721367 | 0.08 |
DCN |
decorin |
144391 |
0.05 |
| chr4_86700210_86700446 | 0.08 |
ARHGAP24 |
Rho GTPase activating protein 24 |
469 |
0.88 |
| chr5_15752995_15753164 | 0.07 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
136988 |
0.05 |
| chr15_42749089_42749416 | 0.06 |
ZNF106 |
zinc finger protein 106 |
459 |
0.78 |
| chr17_48274836_48274996 | 0.06 |
COL1A1 |
collagen, type I, alpha 1 |
2636 |
0.17 |
| chr9_6218213_6218406 | 0.06 |
IL33 |
interleukin 33 |
2500 |
0.39 |
| chr9_113530116_113530340 | 0.06 |
MUSK |
muscle, skeletal, receptor tyrosine kinase |
7399 |
0.3 |
| chr18_72214039_72214190 | 0.06 |
CNDP1 |
carnosine dipeptidase 1 (metallopeptidase M20 family) |
12277 |
0.18 |
| chr15_49715580_49715731 | 0.06 |
FGF7 |
fibroblast growth factor 7 |
198 |
0.96 |
| chr2_238318512_238318719 | 0.06 |
COL6A3 |
collagen, type VI, alpha 3 |
4176 |
0.24 |
| chr9_13568681_13569117 | 0.06 |
MPDZ |
multiple PDZ domain protein |
289310 |
0.01 |
| chr8_93115764_93116215 | 0.06 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
475 |
0.9 |
| chr18_73688861_73689088 | 0.06 |
RP11-94B19.4 |
Uncharacterized protein |
282147 |
0.01 |
| chr5_53833650_53833801 | 0.06 |
SNX18 |
sorting nexin 18 |
20132 |
0.26 |
| chr2_72949822_72949973 | 0.06 |
ENSG00000222536 |
. |
962 |
0.65 |
| chr19_30865612_30866061 | 0.05 |
ZNF536 |
zinc finger protein 536 |
2517 |
0.44 |
| chr17_53275147_53275298 | 0.05 |
HLF |
hepatic leukemia factor |
67151 |
0.12 |
| chr3_120215962_120216316 | 0.05 |
FSTL1 |
follistatin-like 1 |
46039 |
0.16 |
| chr6_80927767_80927918 | 0.05 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
111459 |
0.07 |
| chr3_16219029_16219297 | 0.05 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
2947 |
0.33 |
| chr11_65639318_65639911 | 0.05 |
EFEMP2 |
EGF containing fibulin-like extracellular matrix protein 2 |
511 |
0.56 |
| chr2_189839524_189839773 | 0.05 |
COL3A1 |
collagen, type III, alpha 1 |
549 |
0.79 |
| chr7_93465582_93465846 | 0.05 |
TFPI2 |
tissue factor pathway inhibitor 2 |
53768 |
0.12 |
| chr6_81938853_81939008 | 0.05 |
RP1-300G12.2 |
|
305700 |
0.01 |
| chr10_34630198_34630349 | 0.05 |
PARD3 |
par-3 family cell polarity regulator |
31188 |
0.25 |
| chr5_54864914_54865117 | 0.05 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
34137 |
0.18 |
| chr11_26211058_26211209 | 0.05 |
ANO3 |
anoctamin 3 |
304 |
0.95 |
| chr4_71588376_71588537 | 0.05 |
RUFY3 |
RUN and FYVE domain containing 3 |
84 |
0.95 |
| chr11_102702602_102702762 | 0.05 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
6759 |
0.19 |
| chr9_25647898_25648049 | 0.05 |
TUSC1 |
tumor suppressor candidate 1 |
30883 |
0.27 |
| chr7_94868396_94868601 | 0.05 |
AC002429.5 |
|
24854 |
0.19 |
| chr9_109049783_109049934 | 0.05 |
ENSG00000200131 |
. |
392400 |
0.01 |
| chr10_81818789_81818977 | 0.05 |
TMEM254 |
transmembrane protein 254 |
19543 |
0.18 |
| chr4_55095719_55096018 | 0.05 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
142 |
0.98 |
| chr3_114169134_114169310 | 0.05 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
4308 |
0.3 |
| chr2_154892286_154892437 | 0.05 |
GALNT13 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13) |
91350 |
0.09 |
| chr15_62916629_62916780 | 0.05 |
RP11-625H11.1 |
Uncharacterized protein |
20676 |
0.19 |
| chr5_15533409_15533775 | 0.05 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
32045 |
0.23 |
| chr6_46293041_46293192 | 0.05 |
RCAN2 |
regulator of calcineurin 2 |
289 |
0.94 |
| chr1_150245669_150245859 | 0.05 |
C1orf54 |
chromosome 1 open reading frame 54 |
581 |
0.54 |
| chr2_220942198_220942388 | 0.05 |
ENSG00000221199 |
. |
114306 |
0.06 |
| chr16_89990293_89990742 | 0.05 |
TUBB3 |
Tubulin beta-3 chain |
743 |
0.44 |
| chr11_125931766_125932579 | 0.05 |
CDON |
cell adhesion associated, oncogene regulated |
534 |
0.8 |
| chr10_88518871_88519024 | 0.05 |
BMPR1A |
bone morphogenetic protein receptor, type IA |
2540 |
0.23 |
| chr7_93997822_93997995 | 0.05 |
COL1A2 |
collagen, type I, alpha 2 |
25965 |
0.24 |
| chr11_102667530_102667681 | 0.04 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
1286 |
0.4 |
| chr1_201619186_201619405 | 0.04 |
NAV1 |
neuron navigator 1 |
1845 |
0.29 |
| chr4_26219717_26219868 | 0.04 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
54437 |
0.17 |
| chr2_163098757_163099006 | 0.04 |
FAP |
fibroblast activation protein, alpha |
677 |
0.75 |
| chr4_86397582_86397737 | 0.04 |
ARHGAP24 |
Rho GTPase activating protein 24 |
1268 |
0.64 |
| chr11_121970307_121971111 | 0.04 |
ENSG00000207971 |
. |
157 |
0.79 |
| chr2_175711229_175711408 | 0.04 |
CHN1 |
chimerin 1 |
175 |
0.97 |
| chr4_38705158_38705309 | 0.04 |
RP11-617D20.1 |
|
38729 |
0.12 |
| chr18_53530407_53530634 | 0.04 |
TCF4 |
transcription factor 4 |
198502 |
0.03 |
| chr2_230035602_230035855 | 0.04 |
PID1 |
phosphotyrosine interaction domain containing 1 |
61073 |
0.14 |
| chr10_75759595_75759746 | 0.04 |
VCL |
vinculin |
1796 |
0.32 |
| chr1_57038997_57039148 | 0.04 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
6169 |
0.3 |
| chr2_189441923_189442074 | 0.04 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
7188 |
0.33 |
| chr6_109496434_109496585 | 0.04 |
CEP57L1 |
centrosomal protein 57kDa-like 1 |
19515 |
0.21 |
| chr2_198540036_198540213 | 0.04 |
RFTN2 |
raftlin family member 2 |
595 |
0.74 |
| chr11_114003372_114003692 | 0.04 |
ENSG00000221112 |
. |
45881 |
0.15 |
| chr4_174449079_174449230 | 0.04 |
HAND2-AS1 |
HAND2 antisense RNA 1 (head to head) |
597 |
0.73 |
| chr8_108316001_108316205 | 0.04 |
ANGPT1 |
angiopoietin 1 |
32647 |
0.22 |
| chr18_52969588_52969739 | 0.04 |
TCF4 |
transcription factor 4 |
194 |
0.97 |
| chr2_239696394_239696545 | 0.04 |
TWIST2 |
twist family bHLH transcription factor 2 |
60204 |
0.14 |
| chr3_114790362_114790748 | 0.04 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
333 |
0.94 |
| chr21_28337250_28337401 | 0.04 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
1507 |
0.46 |
| chr13_74315255_74315493 | 0.04 |
KLF12 |
Kruppel-like factor 12 |
253812 |
0.02 |
| chr1_67001120_67001322 | 0.04 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
1256 |
0.58 |
| chr18_57362819_57362970 | 0.04 |
RP11-2N1.2 |
|
797 |
0.55 |
| chr4_151600359_151600541 | 0.04 |
ENSG00000221235 |
. |
39432 |
0.19 |
| chr6_121843994_121844420 | 0.04 |
ENSG00000201379 |
. |
19565 |
0.2 |
| chr1_204034501_204034712 | 0.04 |
SOX13 |
SRY (sex determining region Y)-box 13 |
8126 |
0.2 |
| chr4_4881760_4881911 | 0.04 |
MSX1 |
msh homeobox 1 |
20442 |
0.23 |
| chr5_980561_980809 | 0.04 |
RP11-661C8.3 |
Uncharacterized protein |
1205 |
0.39 |
| chr4_17060566_17060738 | 0.04 |
LDB2 |
LIM domain binding 2 |
160220 |
0.04 |
| chr1_79471738_79472138 | 0.04 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
465 |
0.9 |
| chr14_27065632_27065895 | 0.04 |
NOVA1 |
neuro-oncological ventral antigen 1 |
482 |
0.74 |
| chr5_71920063_71920214 | 0.04 |
ZNF366 |
zinc finger protein 366 |
116584 |
0.06 |
| chr10_30184757_30184953 | 0.04 |
SVIL |
supervillin |
160122 |
0.04 |
| chr8_116673723_116673890 | 0.04 |
TRPS1 |
trichorhinophalangeal syndrome I |
99 |
0.98 |
| chr3_66139685_66140118 | 0.04 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
20616 |
0.25 |
| chr5_121462703_121462886 | 0.04 |
ZNF474 |
zinc finger protein 474 |
2414 |
0.29 |
| chr15_49718765_49718970 | 0.04 |
FGF7 |
fibroblast growth factor 7 |
3410 |
0.29 |
| chr6_132437595_132437746 | 0.04 |
ENSG00000265669 |
. |
1267 |
0.6 |
| chr16_62023154_62023305 | 0.04 |
CDH8 |
cadherin 8, type 2 |
32277 |
0.22 |
| chr2_141579747_141579898 | 0.04 |
ENSG00000201892 |
. |
170290 |
0.04 |
| chr6_55782858_55783009 | 0.04 |
BMP5 |
bone morphogenetic protein 5 |
42571 |
0.21 |
| chr11_76621964_76622115 | 0.04 |
ENSG00000212030 |
. |
32763 |
0.15 |
| chr11_46670801_46671004 | 0.04 |
ATG13 |
autophagy related 13 |
10070 |
0.15 |
| chr15_49715834_49715985 | 0.04 |
FGF7 |
fibroblast growth factor 7 |
452 |
0.86 |
| chr7_82071524_82071810 | 0.04 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
1364 |
0.61 |
| chr6_141785028_141785179 | 0.04 |
ENSG00000222764 |
. |
22446 |
0.29 |
| chr16_80180711_80180962 | 0.04 |
DYNLRB2 |
dynein, light chain, roadblock-type 2 |
393795 |
0.01 |
| chr3_192575065_192575216 | 0.04 |
MB21D2 |
Mab-21 domain containing 2 |
60810 |
0.14 |
| chr10_52761361_52761512 | 0.04 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
10318 |
0.26 |
| chr19_50006575_50006747 | 0.04 |
ENSG00000207782 |
. |
2536 |
0.08 |
| chr1_119527782_119527933 | 0.04 |
TBX15 |
T-box 15 |
2571 |
0.38 |
| chr5_53752275_53752426 | 0.04 |
HSPB3 |
heat shock 27kDa protein 3 |
905 |
0.7 |
| chr2_176279380_176279680 | 0.04 |
ENSG00000221347 |
. |
84429 |
0.1 |
| chr1_92345537_92345688 | 0.04 |
TGFBR3 |
transforming growth factor, beta receptor III |
6054 |
0.24 |
| chr8_93029441_93029592 | 0.04 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
75 |
0.98 |
| chr9_98412403_98412603 | 0.04 |
DKFZP434H0512 |
Protein LOC100506667; Putative uncharacterized protein DKFZp434H0512 |
122102 |
0.05 |
| chr6_76044253_76044525 | 0.04 |
RP11-415D17.3 |
|
22541 |
0.16 |
| chr9_16795062_16795213 | 0.04 |
BNC2 |
basonuclin 2 |
37149 |
0.2 |
| chr9_6355936_6356087 | 0.04 |
TPD52L3 |
tumor protein D52-like 3 |
27607 |
0.19 |
| chr22_44438594_44438745 | 0.04 |
PARVB |
parvin, beta |
11424 |
0.24 |
| chr2_189158808_189158959 | 0.04 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
338 |
0.89 |
| chr5_73944879_73945040 | 0.04 |
ENC1 |
ectodermal-neural cortex 1 (with BTB domain) |
7710 |
0.21 |
| chr10_102278895_102279852 | 0.04 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
218 |
0.92 |
| chr9_99277013_99277426 | 0.04 |
CDC14B |
cell division cycle 14B |
51893 |
0.13 |
| chr8_70180166_70180390 | 0.04 |
ENSG00000238808 |
. |
157469 |
0.04 |
| chr9_89410402_89410553 | 0.04 |
GAS1 |
growth arrest-specific 1 |
151627 |
0.04 |
| chr13_52917687_52917849 | 0.04 |
TPTE2P2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 pseudogene 2 |
45805 |
0.13 |
| chr6_20680927_20681087 | 0.04 |
RP3-348I23.2 |
|
119918 |
0.06 |
| chr8_69905056_69905287 | 0.04 |
ENSG00000238808 |
. |
117638 |
0.07 |
| chr1_67244592_67244787 | 0.04 |
INSL5 |
insulin-like 5 |
22250 |
0.15 |
| chr17_78044749_78045392 | 0.04 |
CCDC40 |
coiled-coil domain containing 40 |
12346 |
0.15 |
| chr11_27404858_27405009 | 0.04 |
CCDC34 |
coiled-coil domain containing 34 |
19518 |
0.22 |
| chr18_66469346_66469497 | 0.04 |
CCDC102B |
coiled-coil domain containing 102B |
3769 |
0.28 |
| chr5_148787044_148787195 | 0.04 |
ENSG00000208035 |
. |
21362 |
0.11 |
| chr8_42054040_42054280 | 0.04 |
PLAT |
plasminogen activator, tissue |
944 |
0.52 |
| chr8_23738839_23738990 | 0.04 |
STC1 |
stanniocalcin 1 |
26594 |
0.21 |
| chr6_44510355_44510599 | 0.04 |
ENSG00000266619 |
. |
107099 |
0.06 |
| chr10_94725160_94725374 | 0.04 |
EXOC6 |
exocyst complex component 6 |
31964 |
0.2 |
| chr15_89181571_89183025 | 0.04 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
114 |
0.96 |
| chr2_178498869_178499062 | 0.04 |
TTC30A |
tetratricopeptide repeat domain 30A |
15271 |
0.18 |
| chr1_172109829_172110023 | 0.04 |
ENSG00000207949 |
. |
1879 |
0.31 |
| chr13_24807615_24808021 | 0.04 |
SPATA13 |
spermatogenesis associated 13 |
18014 |
0.17 |
| chr15_25915468_25915755 | 0.04 |
ATP10A |
ATPase, class V, type 10A |
24361 |
0.19 |
| chr4_173978999_173979247 | 0.04 |
ENSG00000241652 |
. |
83671 |
0.09 |
| chr4_54564515_54564666 | 0.04 |
RP11-317M11.1 |
|
2522 |
0.3 |
| chr6_157297379_157297598 | 0.04 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
74981 |
0.12 |
| chr5_153882501_153882652 | 0.04 |
HAND1 |
heart and neural crest derivatives expressed 1 |
24752 |
0.17 |
| chr7_93954375_93954526 | 0.04 |
COL1A2 |
collagen, type I, alpha 2 |
69423 |
0.13 |
| chr5_158438402_158438767 | 0.04 |
CTD-2363C16.2 |
|
25770 |
0.2 |
| chr4_124776288_124776439 | 0.04 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
455240 |
0.01 |
| chr5_158203015_158203166 | 0.04 |
CTD-2363C16.1 |
|
206924 |
0.02 |
| chr14_38371112_38371263 | 0.04 |
LINC00517 |
long intergenic non-protein coding RNA 517 |
142 |
0.97 |
| chr17_38601617_38601849 | 0.04 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
2020 |
0.24 |
| chr7_137675744_137675914 | 0.04 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
10964 |
0.2 |
| chr12_119617823_119618008 | 0.04 |
HSPB8 |
heat shock 22kDa protein 8 |
520 |
0.73 |
| chrX_14130141_14130292 | 0.04 |
ENSG00000264331 |
. |
36206 |
0.21 |
| chr5_125337918_125338211 | 0.04 |
ENSG00000265637 |
. |
194464 |
0.03 |
| chr4_113820259_113820410 | 0.04 |
RP11-119H12.6 |
|
16848 |
0.24 |
| chr12_27761873_27762033 | 0.04 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
9672 |
0.22 |
| chr15_40111813_40111964 | 0.04 |
RP11-37C7.2 |
|
4258 |
0.19 |
| chr5_98107570_98107721 | 0.04 |
RGMB-AS1 |
RGMB antisense RNA 1 |
837 |
0.59 |
| chr4_55063687_55063852 | 0.04 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
31495 |
0.19 |
| chr12_65287703_65287933 | 0.03 |
ENSG00000221564 |
. |
44985 |
0.15 |
| chr8_124037767_124037931 | 0.03 |
DERL1 |
derlin 1 |
41 |
0.97 |
| chr4_178244162_178244313 | 0.03 |
NEIL3 |
nei endonuclease VIII-like 3 (E. coli) |
13247 |
0.21 |
| chr1_78959715_78959866 | 0.03 |
PTGFR |
prostaglandin F receptor (FP) |
3033 |
0.36 |
| chr15_71626398_71626549 | 0.03 |
RP11-592N21.2 |
|
8190 |
0.3 |
| chr1_211813087_211813261 | 0.03 |
ENSG00000222080 |
. |
13717 |
0.17 |
| chr17_66594592_66594790 | 0.03 |
FAM20A |
family with sequence similarity 20, member A |
2839 |
0.32 |
| chr9_12794012_12794163 | 0.03 |
LURAP1L |
leucine rich adaptor protein 1-like |
19067 |
0.18 |
| chr6_169558527_169558678 | 0.03 |
XXyac-YX65C7_A.2 |
|
54747 |
0.16 |
| chr10_32096317_32096468 | 0.03 |
ARHGAP12 |
Rho GTPase activating protein 12 |
46716 |
0.16 |
| chr15_73263869_73264224 | 0.03 |
NEO1 |
neogenin 1 |
80005 |
0.1 |
| chr22_36651230_36651518 | 0.03 |
APOL1 |
apolipoprotein L, 1 |
2167 |
0.29 |
| chr1_162605279_162605430 | 0.03 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
3094 |
0.27 |
| chr1_25525492_25525809 | 0.03 |
RP4-706G24.1 |
|
8980 |
0.19 |
| chrX_114467670_114468385 | 0.03 |
LRCH2 |
leucine-rich repeats and calponin homology (CH) domain containing 2 |
601 |
0.77 |
| chr12_54398539_54398690 | 0.03 |
HOXC8 |
homeobox C8 |
4218 |
0.07 |
| chr1_78960116_78960267 | 0.03 |
PTGFR |
prostaglandin F receptor (FP) |
3434 |
0.34 |
| chr12_91570586_91570804 | 0.03 |
DCN |
decorin |
1634 |
0.5 |
| chr1_79154291_79154442 | 0.03 |
ENSG00000221683 |
. |
1621 |
0.44 |
| chr12_20526752_20526903 | 0.03 |
RP11-284H19.1 |
|
3631 |
0.29 |
| chr22_29489451_29489602 | 0.03 |
CTA-747E2.10 |
|
18145 |
0.13 |
| chr18_34273309_34273594 | 0.03 |
FHOD3 |
formin homology 2 domain containing 3 |
25069 |
0.22 |
| chr3_120149251_120149402 | 0.03 |
FSTL1 |
follistatin-like 1 |
20512 |
0.22 |
| chr3_106064913_106065064 | 0.03 |
ENSG00000200610 |
. |
169756 |
0.04 |
| chr3_11849089_11849240 | 0.03 |
TAMM41 |
TAM41, mitochondrial translocator assembly and maintenance protein, homolog (S. cerevisiae) |
39082 |
0.19 |
| chr2_153210875_153211026 | 0.03 |
FMNL2 |
formin-like 2 |
19199 |
0.26 |
| chr10_15390844_15391029 | 0.03 |
FAM171A1 |
family with sequence similarity 171, member A1 |
22122 |
0.25 |
| chr17_1665908_1666244 | 0.03 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
47 |
0.96 |
| chr12_59414349_59414500 | 0.03 |
RP11-150C16.1 |
|
100004 |
0.08 |
| chr18_32290517_32290846 | 0.03 |
DTNA |
dystrobrevin, alpha |
420 |
0.9 |
| chr12_53443889_53444561 | 0.03 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
255 |
0.86 |
| chr13_33627122_33627355 | 0.03 |
ENSG00000221677 |
. |
11749 |
0.24 |
| chr9_108059451_108059602 | 0.03 |
SLC44A1 |
solute carrier family 44 (choline transporter), member 1 |
52623 |
0.17 |
| chr7_15554887_15555236 | 0.03 |
AGMO |
alkylglycerol monooxygenase |
46579 |
0.2 |
| chr1_119524620_119524878 | 0.03 |
TBX15 |
T-box 15 |
5679 |
0.29 |
| chr13_80657839_80658035 | 0.03 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
255857 |
0.02 |
| chr15_31528566_31528785 | 0.03 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
75199 |
0.1 |
| chr14_35181195_35181456 | 0.03 |
CFL2 |
cofilin 2 (muscle) |
1709 |
0.38 |
| chr7_43289288_43289439 | 0.03 |
AC004692.4 |
|
496 |
0.82 |
| chr1_92346928_92347079 | 0.03 |
TGFBR3 |
transforming growth factor, beta receptor III |
4663 |
0.25 |
| chr2_177356582_177356733 | 0.03 |
ENSG00000221304 |
. |
108353 |
0.06 |
| chr13_36047635_36048256 | 0.03 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
2887 |
0.26 |
| chr13_36931940_36932091 | 0.03 |
ENSG00000266047 |
. |
2087 |
0.25 |
| chr2_46946794_46946945 | 0.03 |
SOCS5 |
suppressor of cytokine signaling 5 |
20543 |
0.19 |
| chr15_61326560_61326811 | 0.03 |
RP11-39M21.1 |
|
146763 |
0.04 |
| chr2_225847014_225847203 | 0.03 |
ENSG00000263828 |
. |
28149 |
0.22 |
| chr10_14051355_14052213 | 0.03 |
FRMD4A |
FERM domain containing 4A |
1252 |
0.51 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.0 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |