Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
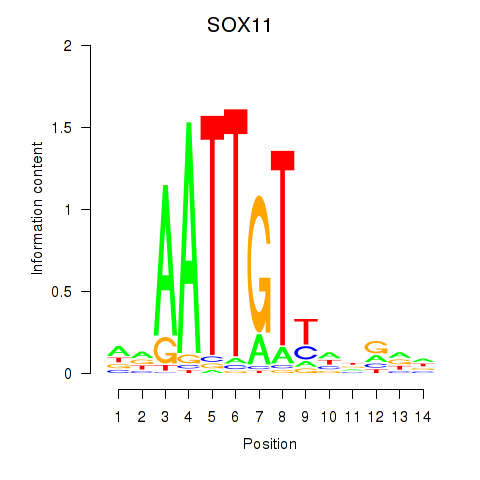
Results for SOX11
Z-value: 1.08
Transcription factors associated with SOX11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX11
|
ENSG00000176887.5 | SRY-box transcription factor 11 |
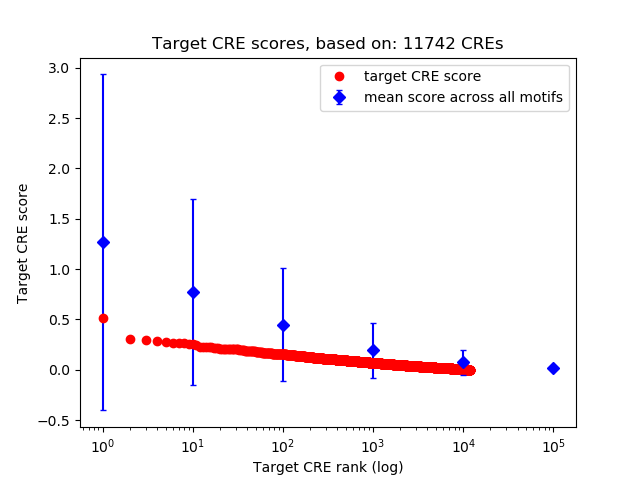
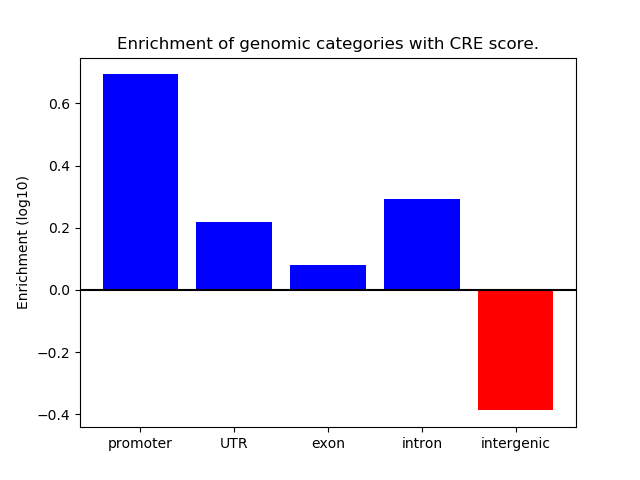
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_5837836_5837987 | SOX11 | 5112 | 0.215759 | -0.51 | 1.6e-01 | Click! |
| chr2_5866291_5866487 | SOX11 | 33590 | 0.169456 | -0.37 | 3.3e-01 | Click! |
| chr2_5835655_5835987 | SOX11 | 3022 | 0.248581 | -0.31 | 4.2e-01 | Click! |
| chr2_5836055_5836258 | SOX11 | 3357 | 0.239357 | -0.27 | 4.9e-01 | Click! |
| chr2_5866537_5866688 | SOX11 | 33813 | 0.169102 | -0.25 | 5.1e-01 | Click! |
Activity of the SOX11 motif across conditions
Conditions sorted by the z-value of the SOX11 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
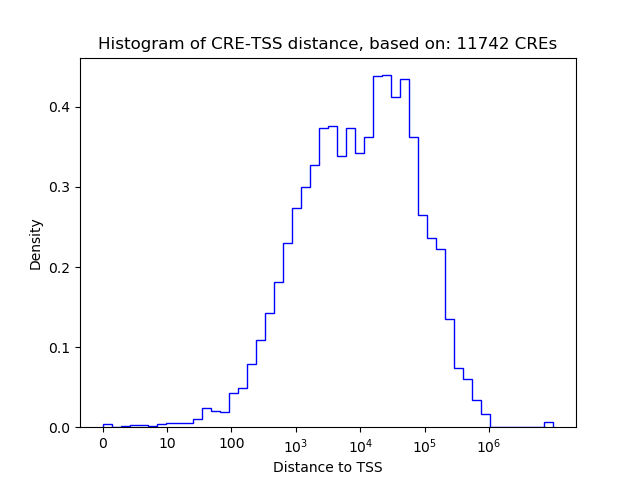
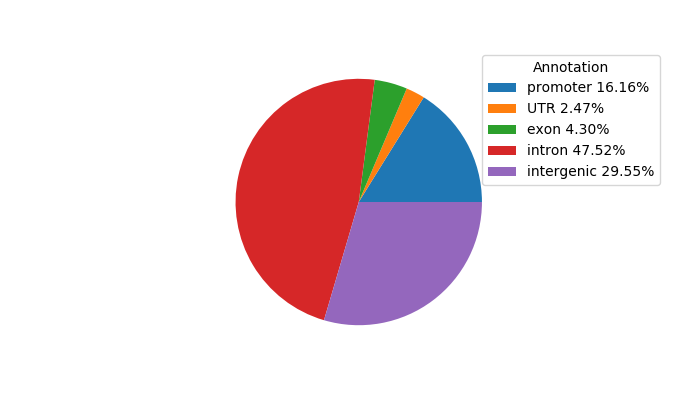
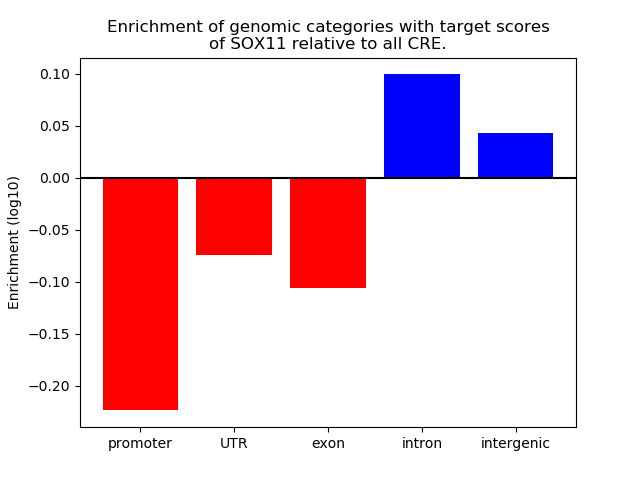
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_160876411_160876699 | 0.51 |
LPAL2 |
lipoprotein, Lp(a)-like 2, pseudogene |
55484 |
0.15 |
| chr12_91572648_91572799 | 0.30 |
DCN |
decorin |
394 |
0.91 |
| chr18_66465806_66465957 | 0.29 |
CCDC102B |
coiled-coil domain containing 102B |
229 |
0.95 |
| chr3_71678241_71678392 | 0.29 |
FOXP1 |
forkhead box P1 |
45176 |
0.15 |
| chr3_178735001_178735152 | 0.28 |
ZMAT3 |
zinc finger, matrin-type 3 |
54278 |
0.14 |
| chr9_115579394_115579717 | 0.27 |
SNX30 |
sorting nexin family member 30 |
21310 |
0.19 |
| chr4_44915041_44915192 | 0.26 |
GNPDA2 |
glucosamine-6-phosphate deaminase 2 |
186504 |
0.03 |
| chr5_42773072_42773223 | 0.26 |
CCDC152 |
coiled-coil domain containing 152 |
16227 |
0.2 |
| chr1_82271501_82271652 | 0.26 |
LPHN2 |
latrophilin 2 |
5494 |
0.35 |
| chr6_135043489_135043640 | 0.25 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
206735 |
0.02 |
| chr2_242318480_242318631 | 0.25 |
FARP2 |
FERM, RhoGEF and pleckstrin domain protein 2 |
6291 |
0.17 |
| chr1_111212990_111213454 | 0.23 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
4433 |
0.21 |
| chr7_43605292_43605443 | 0.23 |
ENSG00000252308 |
. |
13120 |
0.18 |
| chr15_58920203_58920354 | 0.23 |
ENSG00000239100 |
. |
27844 |
0.16 |
| chr4_81195528_81195918 | 0.23 |
FGF5 |
fibroblast growth factor 5 |
7930 |
0.26 |
| chr18_10404815_10404966 | 0.22 |
ENSG00000239031 |
. |
14893 |
0.25 |
| chr12_3001234_3001385 | 0.22 |
TULP3 |
tubby like protein 3 |
1236 |
0.31 |
| chr7_68830446_68830597 | 0.21 |
ENSG00000252423 |
. |
35768 |
0.22 |
| chr17_70372866_70373017 | 0.21 |
SOX9 |
SRY (sex determining region Y)-box 9 |
255780 |
0.02 |
| chr5_148726870_148727055 | 0.21 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
1897 |
0.22 |
| chr2_44625797_44625948 | 0.21 |
CAMKMT |
calmodulin-lysine N-methyltransferase |
25991 |
0.2 |
| chr3_82827750_82827901 | 0.21 |
NA |
NA |
> 106 |
NA |
| chr13_74636535_74636686 | 0.21 |
KLF12 |
Kruppel-like factor 12 |
67424 |
0.15 |
| chr11_4119066_4119217 | 0.21 |
RRM1 |
ribonucleotide reductase M1 |
3068 |
0.26 |
| chr5_88169037_88169242 | 0.21 |
MEF2C |
myocyte enhancer factor 2C |
4655 |
0.27 |
| chr5_156610422_156610951 | 0.21 |
ITK |
IL2-inducible T-cell kinase |
2849 |
0.18 |
| chr15_49719079_49719362 | 0.21 |
FGF7 |
fibroblast growth factor 7 |
3763 |
0.28 |
| chr12_104888025_104888586 | 0.21 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
37526 |
0.2 |
| chrX_13680096_13680380 | 0.20 |
TCEANC |
transcription elongation factor A (SII) N-terminal and central domain containing |
6824 |
0.2 |
| chr2_238777385_238777544 | 0.20 |
ENSG00000263723 |
. |
1083 |
0.53 |
| chr15_80361249_80361400 | 0.20 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
3582 |
0.28 |
| chr2_9736356_9736507 | 0.20 |
ENSG00000207086 |
. |
18965 |
0.16 |
| chr7_93924978_93925129 | 0.20 |
COL1A2 |
collagen, type I, alpha 2 |
98820 |
0.08 |
| chr4_129759342_129759493 | 0.19 |
JADE1 |
jade family PHD finger 1 |
6829 |
0.31 |
| chr3_16929685_16929836 | 0.19 |
PLCL2 |
phospholipase C-like 2 |
3308 |
0.33 |
| chr1_170760279_170760430 | 0.19 |
PRRX1 |
paired related homeobox 1 |
127276 |
0.05 |
| chr14_45711437_45711895 | 0.19 |
MIS18BP1 |
MIS18 binding protein 1 |
10714 |
0.2 |
| chr4_144831533_144831721 | 0.19 |
GYPE |
glycophorin E (MNS blood group) |
4911 |
0.28 |
| chr16_14282323_14282474 | 0.19 |
MKL2 |
MKL/myocardin-like 2 |
1626 |
0.41 |
| chr2_231989300_231989451 | 0.19 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
457 |
0.79 |
| chr1_172109829_172110023 | 0.19 |
ENSG00000207949 |
. |
1879 |
0.31 |
| chr10_95813054_95813205 | 0.19 |
ENSG00000252895 |
. |
6247 |
0.2 |
| chr12_91576386_91576586 | 0.19 |
DCN |
decorin |
0 |
0.99 |
| chr2_134484497_134484648 | 0.19 |
ENSG00000200708 |
. |
130910 |
0.05 |
| chr1_115809994_115810145 | 0.18 |
RP4-663N10.1 |
|
15586 |
0.26 |
| chr10_14630296_14630595 | 0.18 |
FAM107B |
family with sequence similarity 107, member B |
15943 |
0.22 |
| chr8_17554451_17554629 | 0.18 |
MTUS1 |
microtubule associated tumor suppressor 1 |
639 |
0.69 |
| chr15_58900274_58900425 | 0.18 |
ENSG00000239100 |
. |
7915 |
0.2 |
| chr7_15554887_15555236 | 0.18 |
AGMO |
alkylglycerol monooxygenase |
46579 |
0.2 |
| chr7_80178912_80179063 | 0.18 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
37651 |
0.19 |
| chr4_144832724_144832997 | 0.18 |
GYPE |
glycophorin E (MNS blood group) |
6144 |
0.27 |
| chr14_45719805_45719966 | 0.18 |
MIS18BP1 |
MIS18 binding protein 1 |
2495 |
0.3 |
| chr7_81915629_81915780 | 0.17 |
ENSG00000221262 |
. |
3689 |
0.38 |
| chr1_186248731_186248882 | 0.17 |
PRG4 |
proteoglycan 4 |
16599 |
0.19 |
| chr6_107814031_107814182 | 0.17 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
2944 |
0.28 |
| chr11_76621964_76622115 | 0.17 |
ENSG00000212030 |
. |
32763 |
0.15 |
| chr7_90676285_90676436 | 0.17 |
FZD1 |
frizzled family receptor 1 |
217423 |
0.02 |
| chr8_116676008_116676159 | 0.17 |
TRPS1 |
trichorhinophalangeal syndrome I |
2178 |
0.48 |
| chr4_90224084_90224235 | 0.17 |
GPRIN3 |
GPRIN family member 3 |
5002 |
0.34 |
| chr2_118560616_118560767 | 0.17 |
AC009312.1 |
|
1059 |
0.57 |
| chr6_138994650_138994868 | 0.17 |
RP11-390P2.4 |
|
18926 |
0.2 |
| chr12_88966909_88967060 | 0.17 |
KITLG |
KIT ligand |
7254 |
0.25 |
| chr6_81975145_81975342 | 0.17 |
RP1-300G12.2 |
|
269387 |
0.02 |
| chr3_16938891_16939042 | 0.17 |
PLCL2 |
phospholipase C-like 2 |
12514 |
0.25 |
| chr2_204328564_204328715 | 0.17 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
31437 |
0.21 |
| chr4_81170515_81170725 | 0.17 |
FGF5 |
fibroblast growth factor 5 |
17133 |
0.22 |
| chr6_42416461_42416719 | 0.17 |
TRERF1 |
transcriptional regulating factor 1 |
2409 |
0.31 |
| chr5_121509459_121509610 | 0.17 |
CTC-441N14.1 |
|
17824 |
0.18 |
| chr1_147013251_147013702 | 0.16 |
BCL9 |
B-cell CLL/lymphoma 9 |
294 |
0.94 |
| chr17_67412035_67412186 | 0.16 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
1271 |
0.56 |
| chr15_71504545_71504696 | 0.16 |
RP11-673C5.2 |
|
23239 |
0.22 |
| chrX_114276862_114277013 | 0.16 |
ENSG00000222122 |
. |
9947 |
0.21 |
| chr3_159628787_159629360 | 0.16 |
SCHIP1 |
schwannomin interacting protein 1 |
58345 |
0.11 |
| chr11_116915783_116915934 | 0.16 |
ENSG00000200537 |
. |
29245 |
0.13 |
| chr3_60062649_60062918 | 0.16 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
105200 |
0.08 |
| chr12_15283733_15283884 | 0.16 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
7289 |
0.23 |
| chr7_15722890_15723041 | 0.16 |
MEOX2 |
mesenchyme homeobox 2 |
3472 |
0.3 |
| chr18_61647268_61647463 | 0.16 |
SERPINB8 |
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
323 |
0.9 |
| chr4_158973077_158973228 | 0.16 |
FAM198B |
family with sequence similarity 198, member B |
107681 |
0.07 |
| chr6_153321108_153321259 | 0.16 |
MTRF1L |
mitochondrial translational release factor 1-like |
2637 |
0.24 |
| chr3_191008696_191008913 | 0.16 |
UTS2B |
urotensin 2B |
8612 |
0.23 |
| chr4_135976985_135977136 | 0.16 |
ENSG00000207188 |
. |
259617 |
0.02 |
| chr7_19159987_19160138 | 0.16 |
AC003986.7 |
|
506 |
0.73 |
| chr7_15600863_15601014 | 0.16 |
AGMO |
alkylglycerol monooxygenase |
702 |
0.8 |
| chr6_134028778_134028929 | 0.16 |
RP3-323P13.2 |
|
59893 |
0.15 |
| chr5_8686642_8686793 | 0.16 |
ENSG00000247516 |
. |
225679 |
0.02 |
| chr6_135518740_135518891 | 0.16 |
MYB-AS1 |
MYB antisense RNA 1 |
1682 |
0.35 |
| chr6_37410804_37411085 | 0.16 |
CMTR1 |
cap methyltransferase 1 |
9948 |
0.21 |
| chr13_80881448_80881599 | 0.16 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
32271 |
0.23 |
| chr12_28543895_28544046 | 0.16 |
CCDC91 |
coiled-coil domain containing 91 |
59361 |
0.14 |
| chr5_97761082_97761233 | 0.16 |
ENSG00000223053 |
. |
213778 |
0.02 |
| chr9_3732510_3732720 | 0.16 |
RP11-252M18.3 |
|
142969 |
0.05 |
| chr13_74807691_74807851 | 0.16 |
ENSG00000206617 |
. |
55580 |
0.16 |
| chr11_109816867_109817064 | 0.16 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
147122 |
0.05 |
| chr17_46624038_46624312 | 0.16 |
HOXB-AS1 |
HOXB cluster antisense RNA 1 |
121 |
0.85 |
| chr6_157297907_157298141 | 0.16 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
75517 |
0.12 |
| chr1_193506744_193506895 | 0.15 |
ENSG00000252241 |
. |
194255 |
0.03 |
| chr15_31120374_31120525 | 0.15 |
ENSG00000221379 |
. |
26940 |
0.15 |
| chr4_124597002_124597153 | 0.15 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
275954 |
0.02 |
| chr11_63868915_63869066 | 0.15 |
FLRT1 |
fibronectin leucine rich transmembrane protein 1 |
1670 |
0.24 |
| chr3_36504651_36504802 | 0.15 |
STAC |
SH3 and cysteine rich domain |
82661 |
0.11 |
| chr7_13598952_13599103 | 0.15 |
ETV1 |
ets variant 1 |
427039 |
0.01 |
| chr2_236525823_236525974 | 0.15 |
ENSG00000221704 |
. |
45323 |
0.16 |
| chr9_102829140_102829291 | 0.15 |
ERP44 |
endoplasmic reticulum protein 44 |
32107 |
0.16 |
| chr15_25123597_25123815 | 0.15 |
ENSG00000238615 |
. |
15188 |
0.12 |
| chr2_20093347_20093498 | 0.15 |
TTC32 |
tetratricopeptide repeat domain 32 |
7980 |
0.25 |
| chr18_32075255_32075406 | 0.15 |
DTNA |
dystrobrevin, alpha |
1463 |
0.59 |
| chr2_144303459_144303649 | 0.15 |
RP11-570L15.2 |
|
26120 |
0.19 |
| chr2_164202449_164202629 | 0.15 |
ENSG00000200902 |
. |
57034 |
0.18 |
| chr6_26066500_26066651 | 0.15 |
HIST1H1C |
histone cluster 1, H1c |
9876 |
0.07 |
| chr11_9548903_9549060 | 0.15 |
ZNF143 |
zinc finger protein 143 |
14937 |
0.16 |
| chr4_82426507_82426753 | 0.15 |
RASGEF1B |
RasGEF domain family, member 1B |
33561 |
0.24 |
| chr10_53654836_53654987 | 0.15 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
151626 |
0.04 |
| chr4_20447726_20447877 | 0.15 |
SLIT2-IT1 |
SLIT2 intronic transcript 1 (non-protein coding) |
53989 |
0.15 |
| chr7_33947519_33947670 | 0.15 |
BMPER |
BMP binding endothelial regulator |
2449 |
0.44 |
| chr16_22383541_22383722 | 0.15 |
CDR2 |
cerebellar degeneration-related protein 2, 62kDa |
2307 |
0.21 |
| chr19_57684363_57684522 | 0.15 |
DUXA |
double homeobox A |
5631 |
0.13 |
| chr8_66749078_66749602 | 0.15 |
PDE7A |
phosphodiesterase 7A |
1643 |
0.51 |
| chr2_56142145_56142296 | 0.15 |
EFEMP1 |
EGF containing fibulin-like extracellular matrix protein 1 |
8136 |
0.21 |
| chr6_85469897_85470069 | 0.15 |
TBX18 |
T-box 18 |
3090 |
0.38 |
| chrX_135862199_135862350 | 0.15 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
1973 |
0.31 |
| chr18_66917392_66917543 | 0.15 |
DOK6 |
docking protein 6 |
150824 |
0.04 |
| chr12_22486636_22486842 | 0.15 |
ST8SIA1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
358 |
0.93 |
| chr1_200045067_200045218 | 0.15 |
ENSG00000252860 |
. |
21953 |
0.18 |
| chr13_41220599_41220750 | 0.15 |
FOXO1 |
forkhead box O1 |
20060 |
0.22 |
| chr3_108542080_108542294 | 0.14 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
568 |
0.83 |
| chr4_26325438_26325589 | 0.14 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
1021 |
0.7 |
| chr3_16930113_16930264 | 0.14 |
PLCL2 |
phospholipase C-like 2 |
3736 |
0.32 |
| chr5_102841576_102841727 | 0.14 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
56839 |
0.17 |
| chr10_64252438_64252589 | 0.14 |
ZNF365 |
zinc finger protein 365 |
27694 |
0.24 |
| chr12_27769583_27769734 | 0.14 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
17377 |
0.2 |
| chr4_154390309_154390666 | 0.14 |
KIAA0922 |
KIAA0922 |
2986 |
0.33 |
| chrX_130965193_130965374 | 0.14 |
ENSG00000200587 |
. |
107701 |
0.07 |
| chr18_18949042_18949193 | 0.14 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
5563 |
0.25 |
| chr20_30973265_30973471 | 0.14 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
25818 |
0.17 |
| chr7_62828662_62828813 | 0.14 |
AC006455.1 |
|
19498 |
0.21 |
| chr2_177796950_177797101 | 0.14 |
ENSG00000206866 |
. |
2207 |
0.44 |
| chr7_35870864_35871015 | 0.14 |
SEPT7 |
septin 7 |
761 |
0.7 |
| chr2_165658613_165658764 | 0.14 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
28424 |
0.17 |
| chr9_125660522_125660673 | 0.14 |
RC3H2 |
ring finger and CCCH-type domains 2 |
208 |
0.9 |
| chr3_189887899_189888067 | 0.14 |
ENSG00000212489 |
. |
4671 |
0.21 |
| chr2_238317343_238317737 | 0.14 |
COL6A3 |
collagen, type VI, alpha 3 |
5251 |
0.23 |
| chr2_189442201_189442352 | 0.14 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
7466 |
0.32 |
| chr8_100714756_100715301 | 0.14 |
ENSG00000243254 |
. |
88722 |
0.08 |
| chr6_152791599_152791750 | 0.14 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
2091 |
0.37 |
| chr15_77352677_77352828 | 0.14 |
RP11-797A18.6 |
|
7915 |
0.19 |
| chr3_130756724_130756875 | 0.14 |
NEK11 |
NIMA-related kinase 11 |
8246 |
0.16 |
| chr2_65058589_65058850 | 0.14 |
ENSG00000199964 |
. |
2572 |
0.27 |
| chr10_123563818_123563969 | 0.14 |
ENSG00000201884 |
. |
42222 |
0.17 |
| chr4_41198899_41199050 | 0.14 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
17501 |
0.2 |
| chr10_16535131_16535282 | 0.14 |
PTER |
phosphotriesterase related |
8875 |
0.2 |
| chr1_78884908_78885243 | 0.14 |
ENSG00000212308 |
. |
44122 |
0.17 |
| chr12_13307421_13307712 | 0.14 |
EMP1 |
epithelial membrane protein 1 |
42084 |
0.15 |
| chr10_24738869_24739102 | 0.14 |
KIAA1217 |
KIAA1217 |
623 |
0.8 |
| chr6_16671315_16671488 | 0.14 |
RP1-151F17.1 |
|
89968 |
0.09 |
| chr3_143568633_143568784 | 0.14 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
1335 |
0.61 |
| chr5_94891658_94891809 | 0.14 |
ARSK |
arylsulfatase family, member K |
862 |
0.43 |
| chr21_37509990_37510141 | 0.14 |
CBR3 |
carbonyl reductase 3 |
2855 |
0.17 |
| chr1_61559773_61559924 | 0.14 |
NFIA |
nuclear factor I/A |
5764 |
0.22 |
| chr17_1297243_1297394 | 0.14 |
YWHAE |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon |
6157 |
0.19 |
| chr3_59101698_59102091 | 0.14 |
C3orf67 |
chromosome 3 open reading frame 67 |
66084 |
0.15 |
| chr2_109240388_109240570 | 0.14 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
2757 |
0.33 |
| chr15_102354727_102354878 | 0.14 |
OR4F15 |
olfactory receptor, family 4, subfamily F, member 15 |
3564 |
0.18 |
| chr1_236766630_236766781 | 0.13 |
HEATR1 |
HEAT repeat containing 1 |
1083 |
0.54 |
| chr5_8687066_8687217 | 0.13 |
ENSG00000247516 |
. |
226103 |
0.02 |
| chrY_22913829_22913980 | 0.13 |
RPS4Y2 |
ribosomal protein S4, Y-linked 2 |
4146 |
0.37 |
| chr3_192575065_192575216 | 0.13 |
MB21D2 |
Mab-21 domain containing 2 |
60810 |
0.14 |
| chr17_19807297_19807448 | 0.13 |
RP11-209D14.4 |
|
25678 |
0.15 |
| chr5_130882612_130883040 | 0.13 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
14100 |
0.29 |
| chr18_66093771_66093922 | 0.13 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
288448 |
0.01 |
| chr20_12205648_12205799 | 0.13 |
BTBD3 |
BTB (POZ) domain containing 3 |
306224 |
0.01 |
| chr6_159230616_159230767 | 0.13 |
EZR-AS1 |
EZR antisense RNA 1 |
8352 |
0.18 |
| chr7_94027064_94027257 | 0.13 |
COL1A2 |
collagen, type I, alpha 2 |
3287 |
0.35 |
| chr15_49718765_49718970 | 0.13 |
FGF7 |
fibroblast growth factor 7 |
3410 |
0.29 |
| chr6_26569237_26569542 | 0.13 |
ABT1 |
activator of basal transcription 1 |
27791 |
0.13 |
| chr15_56760693_56760949 | 0.13 |
MNS1 |
meiosis-specific nuclear structural 1 |
3486 |
0.34 |
| chr9_79450019_79450170 | 0.13 |
PCA3 |
prostate cancer antigen 3 (non-protein coding) |
70742 |
0.11 |
| chr18_53775687_53775874 | 0.13 |
ENSG00000201816 |
. |
28955 |
0.27 |
| chr1_178105985_178106136 | 0.13 |
RASAL2 |
RAS protein activator like 2 |
42784 |
0.21 |
| chr10_52176430_52176581 | 0.13 |
AC069547.2 |
Uncharacterized protein |
23739 |
0.19 |
| chr2_66806799_66806950 | 0.13 |
MEIS1 |
Meis homeobox 1 |
70815 |
0.12 |
| chr4_16219327_16219478 | 0.13 |
TAPT1 |
transmembrane anterior posterior transformation 1 |
8692 |
0.2 |
| chr16_21657544_21657695 | 0.13 |
ENSG00000207042 |
. |
4004 |
0.14 |
| chr12_80707198_80707349 | 0.13 |
OTOGL |
otogelin-like |
23019 |
0.21 |
| chr4_41198454_41198605 | 0.13 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
17946 |
0.19 |
| chr5_88100959_88101110 | 0.13 |
MEF2C |
myocyte enhancer factor 2C |
18571 |
0.24 |
| chr10_65175846_65175997 | 0.13 |
ENSG00000221063 |
. |
43113 |
0.15 |
| chrY_14900158_14900309 | 0.13 |
USP9Y |
ubiquitin specific peptidase 9, Y-linked |
58737 |
0.16 |
| chr8_113941673_113941824 | 0.13 |
CSMD3 |
CUB and Sushi multiple domains 3 |
239477 |
0.02 |
| chr3_22799861_22800012 | 0.13 |
UBE2E2-AS1 |
UBE2E2 antisense RNA 1 (head to head) |
444088 |
0.01 |
| chr5_124090679_124090914 | 0.13 |
ZNF608 |
zinc finger protein 608 |
6296 |
0.15 |
| chr2_109102010_109102161 | 0.13 |
GCC2 |
GRIP and coiled-coil domain containing 2 |
13784 |
0.21 |
| chr4_120135975_120136177 | 0.13 |
USP53 |
ubiquitin specific peptidase 53 |
2285 |
0.28 |
| chr13_51485798_51485986 | 0.13 |
RNASEH2B-AS1 |
RNASEH2B antisense RNA 1 |
1044 |
0.49 |
| chr2_160996867_160997018 | 0.13 |
ITGB6 |
integrin, beta 6 |
59821 |
0.14 |
| chr1_8360871_8361022 | 0.13 |
ENSG00000251977 |
. |
8623 |
0.16 |
| chr5_116838988_116839139 | 0.13 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
928433 |
0.0 |
| chr15_56533191_56533342 | 0.13 |
RFX7 |
regulatory factor X, 7 |
2198 |
0.3 |
| chr12_100957838_100957989 | 0.13 |
GAS2L3 |
growth arrest-specific 2 like 3 |
9548 |
0.24 |
| chr1_165564678_165564829 | 0.12 |
RP11-280O1.2 |
|
13361 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.3 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.1 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.0 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0002857 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.0 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0015853 | adenine transport(GO:0015853) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0002246 | wound healing involved in inflammatory response(GO:0002246) inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.0 | GO:0090197 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.0 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.0 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.0 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.0 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0032527 | protein exit from endoplasmic reticulum(GO:0032527) |
| 0.0 | 0.0 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0061043 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.0 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0071028 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0002604 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.5 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0033643 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.0 | 0.1 | GO:0031083 | BLOC complex(GO:0031082) BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |