Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
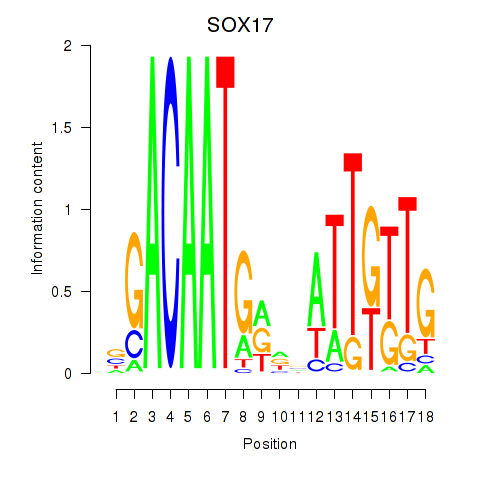
Results for SOX17
Z-value: 1.62
Transcription factors associated with SOX17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX17
|
ENSG00000164736.5 | SRY-box transcription factor 17 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_55379610_55379761 | SOX17 | 9190 | 0.261576 | -0.61 | 8.4e-02 | Click! |
| chr8_55367217_55367368 | SOX17 | 3203 | 0.319968 | -0.60 | 9.0e-02 | Click! |
| chr8_55379904_55380055 | SOX17 | 9484 | 0.260800 | -0.21 | 5.8e-01 | Click! |
| chr8_55382631_55383060 | SOX17 | 12350 | 0.254611 | -0.20 | 6.0e-01 | Click! |
| chr8_55379220_55379441 | SOX17 | 8835 | 0.262574 | 0.16 | 6.9e-01 | Click! |
Activity of the SOX17 motif across conditions
Conditions sorted by the z-value of the SOX17 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
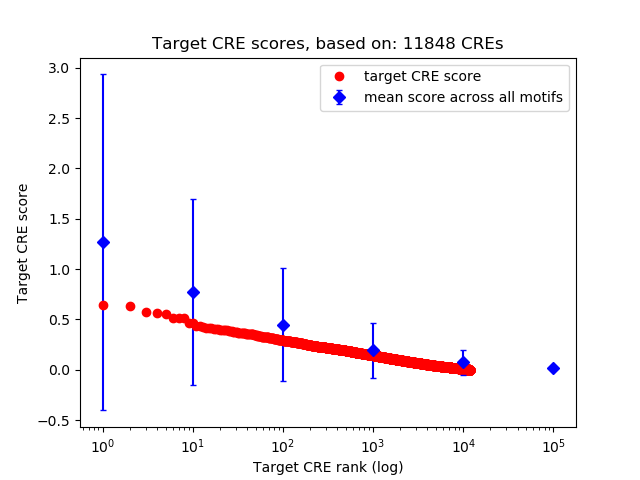
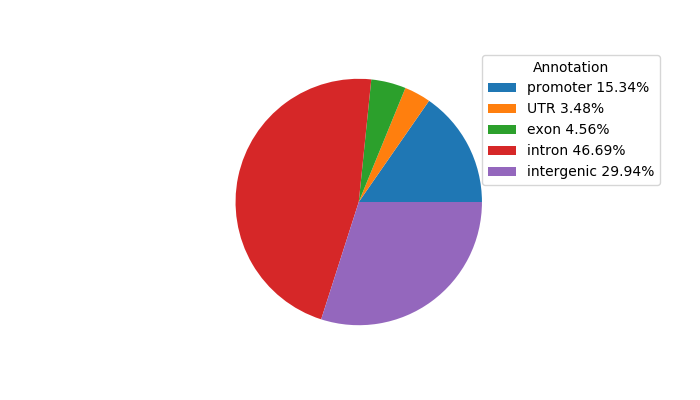
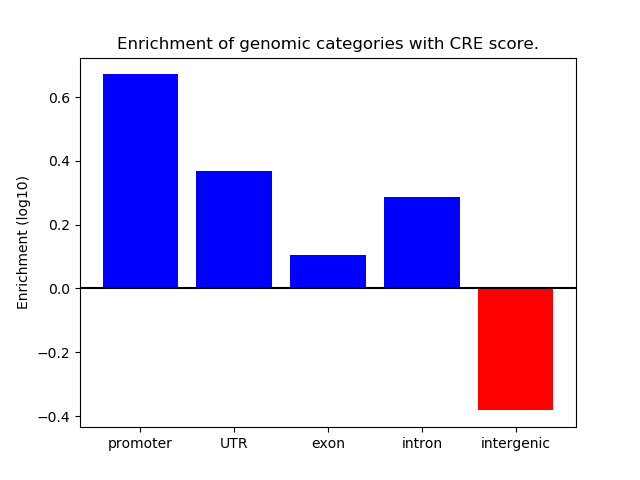
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_39380930_39381081 | 0.65 |
ENSG00000206947 |
. |
12272 |
0.23 |
| chr1_246750885_246751091 | 0.63 |
RP11-439E19.1 |
|
18187 |
0.16 |
| chr7_120630679_120630951 | 0.57 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
1139 |
0.51 |
| chr11_128099186_128099450 | 0.56 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
275971 |
0.01 |
| chr4_181928306_181928668 | 0.56 |
ENSG00000251742 |
. |
827774 |
0.0 |
| chr8_101522130_101522303 | 0.51 |
KB-1615E4.3 |
|
17460 |
0.16 |
| chr4_135119262_135119413 | 0.51 |
PABPC4L |
poly(A) binding protein, cytoplasmic 4-like |
3566 |
0.39 |
| chr22_46454527_46455045 | 0.51 |
RP6-109B7.3 |
|
3991 |
0.12 |
| chr9_21023989_21024197 | 0.47 |
PTPLAD2 |
protein tyrosine phosphatase-like A domain containing 2 |
7515 |
0.22 |
| chr19_34359259_34359976 | 0.47 |
ENSG00000240626 |
. |
58913 |
0.13 |
| chr15_40812177_40812372 | 0.44 |
ENSG00000223313 |
. |
9495 |
0.11 |
| chr17_77817761_77818338 | 0.44 |
CBX4 |
chromobox homolog 4 |
4821 |
0.18 |
| chr6_27205685_27206711 | 0.42 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chr4_682374_683342 | 0.41 |
MFSD7 |
major facilitator superfamily domain containing 7 |
74 |
0.94 |
| chr3_147127121_147127296 | 0.41 |
ZIC1 |
Zic family member 1 |
37 |
0.97 |
| chr2_216299376_216299642 | 0.41 |
FN1 |
fibronectin 1 |
1281 |
0.4 |
| chr2_19849364_19849515 | 0.41 |
TTC32 |
tetratricopeptide repeat domain 32 |
251963 |
0.02 |
| chr7_41740779_41741576 | 0.41 |
INHBA |
inhibin, beta A |
970 |
0.56 |
| chr7_15723235_15723386 | 0.40 |
MEOX2 |
mesenchyme homeobox 2 |
3127 |
0.32 |
| chr16_4161991_4162273 | 0.40 |
ADCY9 |
adenylate cyclase 9 |
1895 |
0.4 |
| chr2_161491624_161491775 | 0.40 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
141394 |
0.05 |
| chr10_17273946_17274204 | 0.39 |
VIM |
vimentin |
1467 |
0.3 |
| chr2_38303069_38303895 | 0.39 |
CYP1B1-AS1 |
CYP1B1 antisense RNA 1 |
156 |
0.5 |
| chr13_24006860_24007105 | 0.39 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
859 |
0.74 |
| chr10_29633011_29633162 | 0.38 |
LYZL1 |
lysozyme-like 1 |
55096 |
0.14 |
| chr3_24527242_24527393 | 0.38 |
ENSG00000228791 |
. |
8261 |
0.19 |
| chr6_148829582_148830289 | 0.38 |
ENSG00000223322 |
. |
15441 |
0.29 |
| chr5_140698139_140698494 | 0.38 |
TAF7 |
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 55kDa |
2014 |
0.13 |
| chr5_127981549_127981700 | 0.38 |
FBN2 |
fibrillin 2 |
13254 |
0.23 |
| chr6_17924518_17924669 | 0.37 |
KIF13A |
kinesin family member 13A |
63101 |
0.13 |
| chr1_65991038_65991651 | 0.37 |
RP4-630A11.3 |
|
39255 |
0.14 |
| chr1_98623502_98623653 | 0.37 |
ENSG00000225206 |
. |
111850 |
0.07 |
| chr7_134469694_134469845 | 0.37 |
CALD1 |
caldesmon 1 |
5340 |
0.32 |
| chr9_18476160_18476529 | 0.37 |
ADAMTSL1 |
ADAMTS-like 1 |
2113 |
0.45 |
| chr15_37395216_37395447 | 0.36 |
MEIS2 |
Meis homeobox 2 |
1827 |
0.38 |
| chr10_3336462_3336663 | 0.36 |
PITRM1 |
pitrilysin metallopeptidase 1 |
121559 |
0.06 |
| chr7_43494536_43494714 | 0.36 |
HECW1 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
24627 |
0.21 |
| chr13_30740237_30740399 | 0.36 |
ENSG00000266816 |
. |
40518 |
0.2 |
| chr5_158176993_158177383 | 0.36 |
CTD-2363C16.1 |
|
232826 |
0.02 |
| chr21_30504177_30504355 | 0.36 |
MAP3K7CL |
MAP3K7 C-terminal like |
688 |
0.61 |
| chr12_111843881_111845308 | 0.36 |
SH2B3 |
SH2B adaptor protein 3 |
842 |
0.62 |
| chr8_93115764_93116215 | 0.36 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
475 |
0.9 |
| chr19_56332953_56333104 | 0.35 |
NLRP11 |
NLR family, pyrin domain containing 11 |
3425 |
0.16 |
| chr16_73087072_73087516 | 0.35 |
ZFHX3 |
zinc finger homeobox 3 |
5020 |
0.25 |
| chr5_151065179_151065606 | 0.35 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
601 |
0.67 |
| chr9_129884150_129884930 | 0.35 |
ANGPTL2 |
angiopoietin-like 2 |
373 |
0.9 |
| chr2_190043861_190044108 | 0.35 |
COL5A2 |
collagen, type V, alpha 2 |
621 |
0.79 |
| chr1_200045338_200045520 | 0.35 |
ENSG00000252860 |
. |
22240 |
0.18 |
| chr4_145570819_145570970 | 0.35 |
HHIP |
hedgehog interacting protein |
3570 |
0.28 |
| chr2_198539523_198539712 | 0.35 |
RFTN2 |
raftlin family member 2 |
1102 |
0.51 |
| chr3_156793670_156793821 | 0.35 |
ENSG00000222499 |
. |
58614 |
0.11 |
| chrX_95691065_95691216 | 0.34 |
ENSG00000223260 |
. |
25359 |
0.28 |
| chr2_145261082_145261233 | 0.34 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
13958 |
0.24 |
| chr18_2856996_2857255 | 0.34 |
EMILIN2 |
elastin microfibril interfacer 2 |
10097 |
0.16 |
| chr21_33893367_33893849 | 0.34 |
ENSG00000252045 |
. |
17001 |
0.16 |
| chr2_150982202_150982420 | 0.33 |
RND3 |
Rho family GTPase 3 |
359585 |
0.01 |
| chr2_69480819_69481174 | 0.33 |
ENSG00000199460 |
. |
70987 |
0.09 |
| chr3_128207705_128209147 | 0.33 |
RP11-475N22.4 |
|
326 |
0.79 |
| chr10_32096471_32096622 | 0.33 |
ARHGAP12 |
Rho GTPase activating protein 12 |
46562 |
0.16 |
| chr10_29824157_29824642 | 0.33 |
ENSG00000207612 |
. |
9627 |
0.19 |
| chr14_56068035_56068254 | 0.33 |
KTN1 |
kinectin 1 (kinesin receptor) |
366 |
0.89 |
| chr3_157158697_157158863 | 0.33 |
PTX3 |
pentraxin 3, long |
4202 |
0.3 |
| chr18_5556544_5556701 | 0.33 |
ENSG00000252576 |
. |
1650 |
0.33 |
| chr4_86702209_86702472 | 0.33 |
ARHGAP24 |
Rho GTPase activating protein 24 |
2481 |
0.39 |
| chr19_46167946_46168097 | 0.32 |
GIPR |
gastric inhibitory polypeptide receptor |
3481 |
0.1 |
| chr8_85513931_85514082 | 0.32 |
ENSG00000206701 |
. |
64094 |
0.14 |
| chr15_96881050_96881298 | 0.32 |
ENSG00000222651 |
. |
4684 |
0.17 |
| chr1_200003972_200004421 | 0.32 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
7185 |
0.18 |
| chr1_101313202_101313363 | 0.32 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
46938 |
0.12 |
| chr4_119763070_119763221 | 0.32 |
SEC24D |
SEC24 family member D |
3320 |
0.33 |
| chr12_19018320_19018530 | 0.32 |
CAPZA3 |
capping protein (actin filament) muscle Z-line, alpha 3 |
127380 |
0.05 |
| chr2_32182398_32182549 | 0.32 |
MEMO1 |
mediator of cell motility 1 |
5894 |
0.21 |
| chr6_106700141_106700423 | 0.32 |
ENSG00000244710 |
. |
31304 |
0.17 |
| chr8_117133093_117133257 | 0.32 |
ENSG00000221793 |
. |
15823 |
0.28 |
| chr11_76621964_76622115 | 0.32 |
ENSG00000212030 |
. |
32763 |
0.15 |
| chr10_25836306_25836457 | 0.31 |
ENSG00000222543 |
. |
153608 |
0.04 |
| chr16_86603613_86603777 | 0.31 |
RP11-463O9.5 |
|
2328 |
0.23 |
| chr1_78960116_78960267 | 0.31 |
PTGFR |
prostaglandin F receptor (FP) |
3434 |
0.34 |
| chr1_31500771_31500983 | 0.31 |
PUM1 |
pumilio RNA-binding family member 1 |
32926 |
0.16 |
| chr4_18059479_18059630 | 0.31 |
LCORL |
ligand dependent nuclear receptor corepressor-like |
36055 |
0.23 |
| chr2_205205743_205205894 | 0.31 |
PARD3B |
par-3 family cell polarity regulator beta |
204698 |
0.03 |
| chr21_36780972_36781123 | 0.31 |
ENSG00000211590 |
. |
311966 |
0.01 |
| chr7_120631344_120631706 | 0.31 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
1849 |
0.35 |
| chr11_112505510_112505661 | 0.31 |
ENSG00000212397 |
. |
32410 |
0.23 |
| chr13_24007256_24007423 | 0.30 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
502 |
0.88 |
| chr15_32963693_32963946 | 0.30 |
RP11-1000B6.2 |
|
1447 |
0.36 |
| chr11_58350202_58350353 | 0.30 |
ZFP91 |
ZFP91 zinc finger protein |
3693 |
0.18 |
| chr1_120189851_120190681 | 0.30 |
ZNF697 |
zinc finger protein 697 |
130 |
0.97 |
| chr3_65583100_65583492 | 0.30 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
114 |
0.98 |
| chr7_83821514_83821694 | 0.30 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
2613 |
0.45 |
| chrX_39949292_39950247 | 0.30 |
BCOR |
BCL6 corepressor |
6887 |
0.33 |
| chr14_53511502_53511653 | 0.30 |
RP11-368P15.3 |
|
8104 |
0.26 |
| chr4_15922747_15922942 | 0.30 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
17127 |
0.2 |
| chr17_9862955_9863106 | 0.30 |
GAS7 |
growth arrest-specific 7 |
251 |
0.94 |
| chr1_95561956_95562107 | 0.30 |
TMEM56 |
transmembrane protein 56 |
20863 |
0.12 |
| chr12_13351879_13352395 | 0.30 |
EMP1 |
epithelial membrane protein 1 |
2417 |
0.37 |
| chr3_25470358_25470509 | 0.29 |
RARB |
retinoic acid receptor, beta |
631 |
0.81 |
| chr6_138864777_138865078 | 0.29 |
NHSL1 |
NHS-like 1 |
1921 |
0.45 |
| chr17_8825496_8825647 | 0.29 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
9737 |
0.23 |
| chr6_155214673_155214881 | 0.29 |
ENSG00000238963 |
. |
11514 |
0.18 |
| chr12_120246427_120246712 | 0.29 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
5382 |
0.27 |
| chr8_102372972_102373210 | 0.29 |
ENSG00000239211 |
. |
37070 |
0.18 |
| chr12_56100408_56100559 | 0.29 |
ITGA7 |
integrin, alpha 7 |
1002 |
0.31 |
| chr6_76460588_76461018 | 0.29 |
MYO6 |
myosin VI |
1754 |
0.36 |
| chr3_21792365_21792565 | 0.29 |
ZNF385D |
zinc finger protein 385D |
462 |
0.89 |
| chr2_12461385_12461536 | 0.29 |
ENSG00000207183 |
. |
90167 |
0.09 |
| chrX_100672866_100673966 | 0.29 |
ENSG00000221132 |
. |
9067 |
0.11 |
| chr6_168048486_168048637 | 0.29 |
AL009178.1 |
Uncharacterized protein; cDNA FLJ43200 fis, clone FEBRA2007793 |
147649 |
0.04 |
| chr7_69470975_69471136 | 0.29 |
AUTS2 |
autism susceptibility candidate 2 |
406458 |
0.01 |
| chr3_157160417_157160653 | 0.29 |
PTX3 |
pentraxin 3, long |
5957 |
0.27 |
| chr5_32505011_32505171 | 0.29 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
26648 |
0.2 |
| chr10_126724055_126724287 | 0.29 |
ENSG00000264572 |
. |
2732 |
0.29 |
| chr5_70315966_70316295 | 0.29 |
NAIP |
NLR family, apoptosis inhibitory protein |
407 |
0.88 |
| chr8_128651347_128651766 | 0.29 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
96124 |
0.08 |
| chr8_83170035_83170244 | 0.29 |
SNX16 |
sorting nexin 16 |
415038 |
0.01 |
| chr6_57369643_57369794 | 0.28 |
ENSG00000212017 |
. |
114788 |
0.07 |
| chr17_46671414_46671766 | 0.28 |
HOXB5 |
homeobox B5 |
267 |
0.75 |
| chr12_13255888_13256039 | 0.28 |
GSG1 |
germ cell associated 1 |
613 |
0.76 |
| chr7_18810301_18810537 | 0.28 |
ENSG00000222164 |
. |
37483 |
0.22 |
| chr10_95239818_95240064 | 0.28 |
MYOF |
myoferlin |
2010 |
0.32 |
| chr6_81232911_81233062 | 0.28 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
416603 |
0.01 |
| chr18_9675042_9675193 | 0.28 |
RAB31 |
RAB31, member RAS oncogene family |
33045 |
0.15 |
| chr3_170147186_170147337 | 0.28 |
CLDN11 |
claudin 11 |
10406 |
0.26 |
| chr12_84472792_84472943 | 0.28 |
ENSG00000221148 |
. |
104360 |
0.09 |
| chr15_96888963_96889465 | 0.28 |
ENSG00000222651 |
. |
12724 |
0.15 |
| chr3_149371362_149371801 | 0.28 |
WWTR1-AS1 |
WWTR1 antisense RNA 1 |
3226 |
0.2 |
| chr2_74413202_74413353 | 0.28 |
CATX-2 |
HCG1989573; Uncharacterized protein |
5240 |
0.14 |
| chr2_54435704_54435855 | 0.28 |
ENSG00000252934 |
. |
41635 |
0.15 |
| chrX_53078746_53079004 | 0.28 |
GPR173 |
G protein-coupled receptor 173 |
410 |
0.85 |
| chr3_32344642_32344793 | 0.28 |
ENSG00000207857 |
. |
43497 |
0.14 |
| chr10_3666400_3666551 | 0.28 |
RP11-184A2.3 |
|
126784 |
0.06 |
| chr10_63818920_63819071 | 0.28 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
10025 |
0.27 |
| chr6_117920563_117920770 | 0.28 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
2861 |
0.34 |
| chr6_3825964_3826270 | 0.28 |
RP11-420L9.4 |
|
6050 |
0.22 |
| chr21_33867197_33867430 | 0.28 |
ENSG00000252045 |
. |
43296 |
0.11 |
| chr8_116675050_116675578 | 0.27 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr10_89822354_89822505 | 0.27 |
ENSG00000200891 |
. |
67977 |
0.12 |
| chr7_92327286_92327437 | 0.27 |
ENSG00000206763 |
. |
3767 |
0.31 |
| chr15_56035451_56035609 | 0.27 |
PRTG |
protogenin |
242 |
0.94 |
| chr4_48256193_48256344 | 0.27 |
TEC |
tec protein tyrosine kinase |
15613 |
0.23 |
| chr13_76213824_76214088 | 0.27 |
LMO7 |
LIM domain 7 |
3497 |
0.21 |
| chr7_55001398_55001578 | 0.27 |
ENSG00000252054 |
. |
67977 |
0.12 |
| chr5_135365829_135365985 | 0.27 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
1216 |
0.52 |
| chr5_74862582_74862812 | 0.27 |
POLK |
polymerase (DNA directed) kappa |
19849 |
0.15 |
| chr12_960802_961189 | 0.27 |
WNK1 |
WNK lysine deficient protein kinase 1 |
136 |
0.97 |
| chr14_73881950_73882101 | 0.27 |
NUMB |
numb homolog (Drosophila) |
5246 |
0.21 |
| chr8_94238484_94238635 | 0.27 |
AC016885.1 |
Uncharacterized protein |
3308 |
0.29 |
| chr12_78336292_78336638 | 0.27 |
NAV3 |
neuron navigator 3 |
23591 |
0.28 |
| chr12_29397730_29397881 | 0.27 |
FAR2 |
fatty acyl CoA reductase 2 |
21127 |
0.23 |
| chr6_11219192_11219343 | 0.27 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
13624 |
0.22 |
| chr11_63647826_63647977 | 0.27 |
ENSG00000202089 |
. |
2256 |
0.22 |
| chr6_46900604_46900792 | 0.27 |
GPR116 |
G protein-coupled receptor 116 |
10984 |
0.22 |
| chr5_172195314_172195493 | 0.27 |
DUSP1 |
dual specificity phosphatase 1 |
2795 |
0.19 |
| chr17_46177753_46179071 | 0.27 |
CBX1 |
chromobox homolog 1 |
148 |
0.91 |
| chr11_10479478_10479629 | 0.27 |
AMPD3 |
adenosine monophosphate deaminase 3 |
1820 |
0.36 |
| chr5_158128189_158128340 | 0.26 |
CTD-2363C16.1 |
|
281750 |
0.01 |
| chr1_186944219_186944581 | 0.26 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
146278 |
0.05 |
| chr12_54017495_54017675 | 0.26 |
ATF7 |
activating transcription factor 7 |
2170 |
0.23 |
| chr9_74295565_74295716 | 0.26 |
TMEM2 |
transmembrane protein 2 |
24208 |
0.27 |
| chr6_18551314_18551465 | 0.26 |
ENSG00000207775 |
. |
20626 |
0.27 |
| chr12_80987527_80987740 | 0.26 |
RP11-272K23.3 |
|
153 |
0.97 |
| chr4_186577993_186578477 | 0.26 |
SORBS2 |
sorbin and SH3 domain containing 2 |
112 |
0.97 |
| chr1_22345507_22345745 | 0.26 |
ENSG00000201273 |
. |
8148 |
0.1 |
| chr6_76504796_76504947 | 0.26 |
MYO6 |
myosin VI |
22369 |
0.17 |
| chr5_60962215_60962366 | 0.26 |
ENSG00000212215 |
. |
2224 |
0.32 |
| chr3_1098193_1098344 | 0.26 |
ENSG00000244169 |
. |
9291 |
0.28 |
| chrX_42106034_42106185 | 0.26 |
ENSG00000206896 |
. |
72471 |
0.12 |
| chr11_133944204_133944355 | 0.26 |
JAM3 |
junctional adhesion molecule 3 |
5313 |
0.26 |
| chr5_121412020_121412268 | 0.26 |
LOX |
lysyl oxidase |
1836 |
0.4 |
| chr15_49715580_49715731 | 0.26 |
FGF7 |
fibroblast growth factor 7 |
198 |
0.96 |
| chr9_91606379_91607009 | 0.26 |
S1PR3 |
sphingosine-1-phosphate receptor 3 |
332 |
0.78 |
| chr8_50969798_50969978 | 0.26 |
SNTG1 |
syntrophin, gamma 1 |
145183 |
0.05 |
| chr13_94361906_94362057 | 0.26 |
GPC6-AS2 |
GPC6 antisense RNA 2 |
126416 |
0.06 |
| chr7_15723868_15724269 | 0.26 |
MEOX2 |
mesenchyme homeobox 2 |
2369 |
0.37 |
| chr20_46043998_46044155 | 0.26 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
58509 |
0.09 |
| chr15_48959812_48960090 | 0.25 |
FBN1 |
fibrillin 1 |
21905 |
0.23 |
| chr11_12204433_12204584 | 0.25 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
20805 |
0.23 |
| chr5_69758456_69758685 | 0.25 |
RP11-497H16.7 |
|
10705 |
0.27 |
| chr3_147648289_147648638 | 0.25 |
ENSG00000221431 |
. |
19175 |
0.29 |
| chr12_115813083_115813371 | 0.25 |
RP11-116D17.1 |
HCG2038717; Uncharacterized protein |
12410 |
0.32 |
| chr21_28168221_28168372 | 0.25 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
47041 |
0.18 |
| chr5_52211157_52211369 | 0.25 |
ITGA2 |
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
73893 |
0.1 |
| chr1_101186076_101186227 | 0.25 |
VCAM1 |
vascular cell adhesion molecule 1 |
831 |
0.73 |
| chr8_97744511_97744662 | 0.25 |
CPQ |
carboxypeptidase Q |
28616 |
0.25 |
| chr8_17814649_17814800 | 0.25 |
PCM1 |
pericentriolar material 1 |
15368 |
0.21 |
| chr10_33230803_33231099 | 0.25 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
5772 |
0.31 |
| chr18_65691492_65691643 | 0.25 |
DSEL |
dermatan sulfate epimerase-like |
507350 |
0.0 |
| chr20_30457693_30458808 | 0.25 |
DUSP15 |
dual specificity phosphatase 15 |
125 |
0.67 |
| chr2_46295244_46295493 | 0.25 |
AC017006.2 |
|
10599 |
0.26 |
| chr4_81192543_81192855 | 0.25 |
FGF5 |
fibroblast growth factor 5 |
4906 |
0.28 |
| chr12_68055164_68055319 | 0.25 |
RP11-335O4.3 |
|
9458 |
0.25 |
| chr4_182878700_182878851 | 0.25 |
ENSG00000251742 |
. |
122514 |
0.06 |
| chr6_11655417_11655591 | 0.25 |
ENSG00000207419 |
. |
54548 |
0.15 |
| chr2_11508011_11508277 | 0.25 |
ROCK2 |
Rho-associated, coiled-coil containing protein kinase 2 |
23433 |
0.2 |
| chr9_20604527_20604679 | 0.25 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
17269 |
0.19 |
| chr18_29239238_29239389 | 0.25 |
ENSG00000252379 |
. |
10028 |
0.16 |
| chr6_17282088_17282659 | 0.25 |
RBM24 |
RNA binding motif protein 24 |
78 |
0.99 |
| chr12_106667138_106667289 | 0.24 |
ENSG00000238609 |
. |
9554 |
0.17 |
| chr17_70339032_70339432 | 0.24 |
SOX9 |
SRY (sex determining region Y)-box 9 |
222071 |
0.02 |
| chr6_19804205_19805055 | 0.24 |
ID4 |
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
32987 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.5 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0052257 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.1 | 0.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.2 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 0.1 | GO:0050942 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 0.2 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.1 | 0.3 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.1 | 0.1 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.3 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.1 | 0.1 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.1 | 0.2 | GO:0014034 | neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) |
| 0.1 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.1 | 0.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.5 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0010800 | regulation of peptidyl-threonine phosphorylation(GO:0010799) positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.2 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.1 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0036296 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.5 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.1 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.0 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.0 | GO:0030910 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.4 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.5 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.0 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.2 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.2 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.0 | GO:0071801 | regulation of podosome assembly(GO:0071801) positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.0 | GO:0060872 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.2 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.0 | 0.1 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.0 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.1 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 | 0.1 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0072338 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0034145 | positive regulation of toll-like receptor 4 signaling pathway(GO:0034145) |
| 0.0 | 0.2 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 0.2 | GO:0050715 | positive regulation of cytokine secretion(GO:0050715) |
| 0.0 | 0.1 | GO:0009820 | alkaloid metabolic process(GO:0009820) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0007442 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.2 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.3 | GO:0070374 | positive regulation of ERK1 and ERK2 cascade(GO:0070374) |
| 0.0 | 0.1 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.6 | GO:0042472 | inner ear morphogenesis(GO:0042472) |
| 0.0 | 0.2 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.2 | GO:0030318 | melanocyte differentiation(GO:0030318) pigment cell differentiation(GO:0050931) |
| 0.0 | 0.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0001865 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.0 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.0 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.5 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.0 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.2 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.1 | GO:0030517 | negative regulation of axon extension(GO:0030517) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.1 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.0 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.0 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.2 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.2 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.9 | GO:0044420 | extracellular matrix component(GO:0044420) |
| 0.0 | 0.1 | GO:0036126 | outer dense fiber(GO:0001520) sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.2 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.1 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.1 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.0 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.5 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.4 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0071617 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) lysophosphatidic acid acyltransferase activity(GO:0042171) lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.0 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.0 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.0 | 0.0 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.0 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 2.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.0 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.6 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |