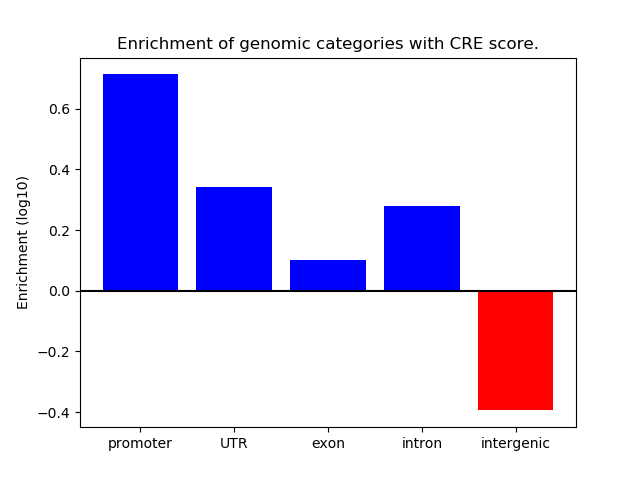
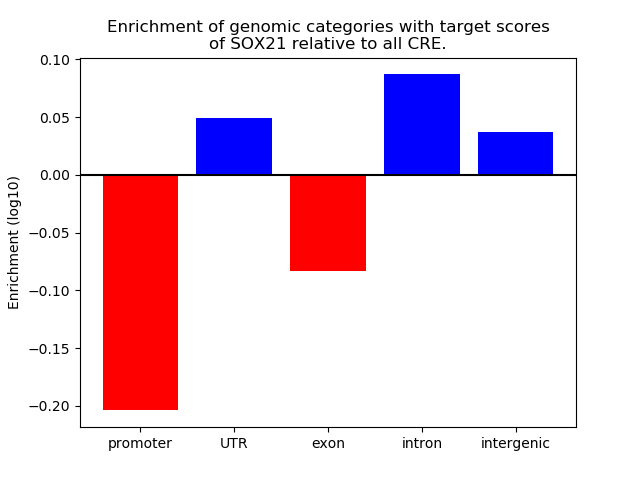
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
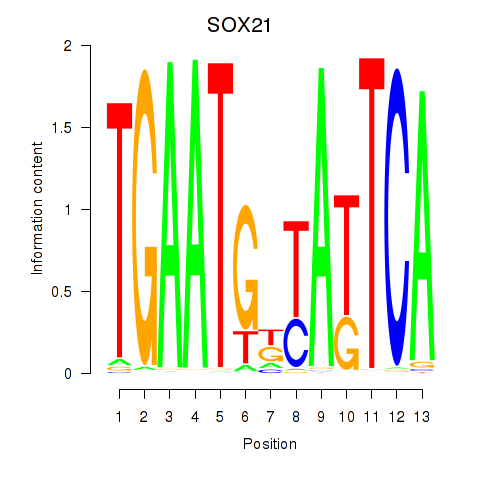
Results for SOX21
Z-value: 0.63
Transcription factors associated with SOX21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX21
|
ENSG00000125285.4 | SRY-box transcription factor 21 |
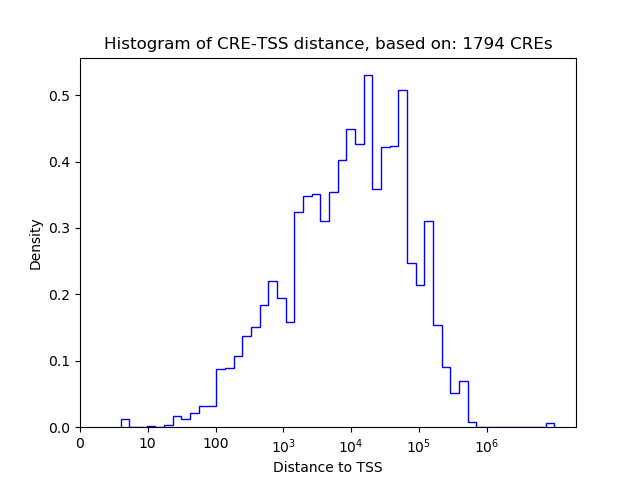
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_95364633_95365589 | SOX21 | 722 | 0.705457 | -0.67 | 4.8e-02 | Click! |
| chr13_95364001_95364370 | SOX21 | 204 | 0.948072 | -0.66 | 5.3e-02 | Click! |
| chr13_95363163_95363594 | SOX21 | 1011 | 0.575324 | -0.57 | 1.1e-01 | Click! |
| chr13_95359649_95360125 | SOX21 | 4502 | 0.232139 | -0.49 | 1.8e-01 | Click! |
| chr13_95364387_95364538 | SOX21 | 73 | 0.976639 | -0.36 | 3.5e-01 | Click! |
Activity of the SOX21 motif across conditions
Conditions sorted by the z-value of the SOX21 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
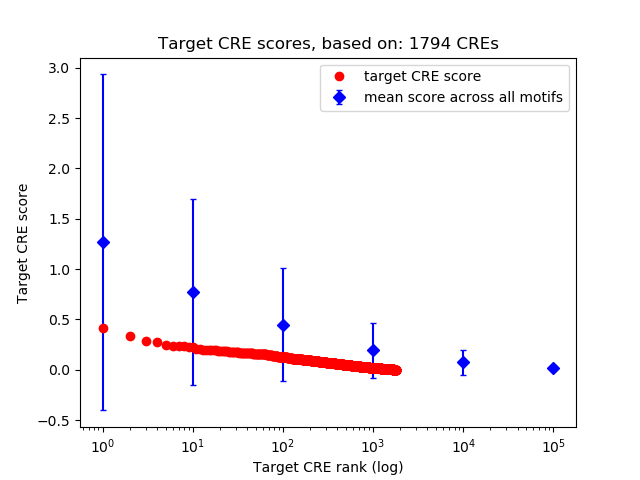
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_63639243_63639445 | 0.42 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
2596 |
0.2 |
| chr18_22655433_22655584 | 0.33 |
ZNF521 |
zinc finger protein 521 |
149245 |
0.05 |
| chr17_15151294_15151445 | 0.28 |
ENSG00000265110 |
. |
3644 |
0.18 |
| chr3_87010891_87011042 | 0.28 |
VGLL3 |
vestigial like 3 (Drosophila) |
28886 |
0.26 |
| chr13_49234522_49234673 | 0.25 |
CYSLTR2 |
cysteinyl leukotriene receptor 2 |
46354 |
0.17 |
| chr6_24858102_24858253 | 0.24 |
ENSG00000263391 |
. |
17884 |
0.17 |
| chr4_135120287_135120438 | 0.24 |
PABPC4L |
poly(A) binding protein, cytoplasmic 4-like |
2541 |
0.45 |
| chr7_137671468_137671947 | 0.23 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
15086 |
0.19 |
| chr6_133073670_133073821 | 0.23 |
RP1-55C23.7 |
|
69 |
0.96 |
| chrX_100545794_100545945 | 0.23 |
TAF7L |
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
455 |
0.73 |
| chr21_39678468_39679041 | 0.21 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
9906 |
0.29 |
| chr6_35570444_35570718 | 0.20 |
ENSG00000212579 |
. |
49014 |
0.11 |
| chr11_128588557_128588803 | 0.20 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
22762 |
0.17 |
| chrX_12971375_12971526 | 0.20 |
TMSB4X |
thymosin beta 4, X-linked |
21777 |
0.18 |
| chrX_108952305_108952710 | 0.20 |
ACSL4 |
acyl-CoA synthetase long-chain family member 4 |
17131 |
0.22 |
| chr13_106828289_106828440 | 0.20 |
ENSG00000222682 |
. |
20525 |
0.25 |
| chrX_70858475_70858626 | 0.20 |
ENSG00000264855 |
. |
12845 |
0.19 |
| chr1_110546930_110547275 | 0.19 |
AHCYL1 |
adenosylhomocysteinase-like 1 |
347 |
0.85 |
| chr12_77081595_77081807 | 0.19 |
ZDHHC17 |
zinc finger, DHHC-type containing 17 |
75667 |
0.11 |
| chr1_92349055_92349206 | 0.19 |
TGFBR3 |
transforming growth factor, beta receptor III |
2536 |
0.32 |
| chr13_63869028_63869179 | 0.18 |
LINC00395 |
long intergenic non-protein coding RNA 395 |
422048 |
0.01 |
| chr14_27067998_27068288 | 0.18 |
NOVA1-AS1 |
NOVA1 antisense RNA 1 (head to head) |
525 |
0.68 |
| chr12_106700881_106701032 | 0.18 |
CKAP4 |
cytoskeleton-associated protein 4 |
2899 |
0.22 |
| chr8_66867885_66868036 | 0.18 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
65835 |
0.13 |
| chr3_73795869_73796020 | 0.18 |
PDZRN3 |
PDZ domain containing ring finger 3 |
121853 |
0.06 |
| chr18_52919364_52919515 | 0.18 |
TCF4 |
transcription factor 4 |
23463 |
0.25 |
| chr11_71847779_71847930 | 0.18 |
FOLR3 |
folate receptor 3 (gamma) |
960 |
0.35 |
| chr14_62180129_62180280 | 0.18 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
15864 |
0.23 |
| chr6_160135650_160135801 | 0.17 |
ENSG00000251988 |
. |
5847 |
0.15 |
| chr1_212794908_212795059 | 0.17 |
FAM71A |
family with sequence similarity 71, member A |
2806 |
0.19 |
| chr16_81852082_81852236 | 0.17 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
39296 |
0.2 |
| chr18_74189003_74189166 | 0.17 |
ZNF516 |
zinc finger protein 516 |
13651 |
0.17 |
| chr2_145273150_145273649 | 0.17 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
1716 |
0.39 |
| chr1_169574529_169574680 | 0.17 |
SELP |
selectin P (granule membrane protein 140kDa, antigen CD62) |
13853 |
0.2 |
| chr8_1954691_1954842 | 0.17 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
32722 |
0.19 |
| chr19_2783530_2783806 | 0.17 |
SGTA |
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
299 |
0.8 |
| chr8_48504664_48504815 | 0.17 |
RP11-697N18.4 |
|
64780 |
0.11 |
| chr4_16030817_16031107 | 0.17 |
ENSG00000251758 |
. |
20552 |
0.19 |
| chr7_28727324_28727475 | 0.17 |
CREB5 |
cAMP responsive element binding protein 5 |
1801 |
0.52 |
| chr5_177628923_177629074 | 0.16 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
2510 |
0.27 |
| chr1_110159560_110159711 | 0.16 |
AMPD2 |
adenosine monophosphate deaminase 2 |
909 |
0.36 |
| chr2_65025688_65025839 | 0.16 |
ENSG00000239891 |
. |
18749 |
0.17 |
| chr5_15510117_15510353 | 0.16 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
8688 |
0.31 |
| chr12_48588557_48588708 | 0.16 |
DKFZP779L1853 |
|
3538 |
0.16 |
| chr5_87441681_87441853 | 0.16 |
TMEM161B |
transmembrane protein 161B |
74681 |
0.12 |
| chr7_37008930_37009081 | 0.16 |
ELMO1 |
engulfment and cell motility 1 |
15660 |
0.2 |
| chr17_65236131_65236424 | 0.16 |
HELZ |
helicase with zinc finger |
361 |
0.82 |
| chr1_110053417_110053568 | 0.16 |
AMIGO1 |
adhesion molecule with Ig-like domain 1 |
1132 |
0.33 |
| chr6_12762324_12762595 | 0.16 |
ENSG00000223291 |
. |
8552 |
0.25 |
| chr2_175711229_175711408 | 0.16 |
CHN1 |
chimerin 1 |
175 |
0.97 |
| chr4_109079309_109079460 | 0.16 |
LEF1 |
lymphoid enhancer-binding factor 1 |
8073 |
0.23 |
| chr1_178055880_178056031 | 0.16 |
RASAL2 |
RAS protein activator like 2 |
6909 |
0.32 |
| chr2_158299517_158299677 | 0.16 |
CYTIP |
cytohesin 1 interacting protein |
1057 |
0.48 |
| chr3_185741709_185741860 | 0.16 |
ENSG00000266670 |
. |
41701 |
0.14 |
| chr3_45957301_45957532 | 0.16 |
LZTFL1 |
leucine zipper transcription factor-like 1 |
118 |
0.95 |
| chr15_39539947_39540098 | 0.15 |
C15orf54 |
chromosome 15 open reading frame 54 |
2863 |
0.34 |
| chr14_35181195_35181456 | 0.15 |
CFL2 |
cofilin 2 (muscle) |
1709 |
0.38 |
| chr1_96483696_96483847 | 0.15 |
RP11-147C23.1 |
Uncharacterized protein |
26226 |
0.26 |
| chr10_32096317_32096468 | 0.15 |
ARHGAP12 |
Rho GTPase activating protein 12 |
46716 |
0.16 |
| chr12_20000514_20000781 | 0.15 |
ENSG00000200885 |
. |
123278 |
0.06 |
| chrX_153773485_153773690 | 0.15 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
555 |
0.53 |
| chr3_145654583_145654734 | 0.15 |
RP11-274H2.5 |
|
124808 |
0.05 |
| chr11_58978296_58978447 | 0.15 |
MPEG1 |
macrophage expressed 1 |
2053 |
0.27 |
| chr6_56010133_56010284 | 0.15 |
COL21A1 |
collagen, type XXI, alpha 1 |
21527 |
0.28 |
| chr5_126745531_126745682 | 0.15 |
PRRC1 |
proline-rich coiled-coil 1 |
107695 |
0.07 |
| chr18_53005046_53005364 | 0.15 |
TCF4 |
transcription factor 4 |
14023 |
0.28 |
| chr6_129819252_129819465 | 0.15 |
RP1-69D17.3 |
|
16803 |
0.26 |
| chr12_46759532_46760006 | 0.15 |
SLC38A2 |
solute carrier family 38, member 2 |
679 |
0.75 |
| chr12_2163640_2163867 | 0.15 |
CACNA1C |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
1024 |
0.49 |
| chr7_81991326_81991606 | 0.15 |
ENSG00000221262 |
. |
72073 |
0.12 |
| chr6_3868889_3869175 | 0.15 |
FAM50B |
family with sequence similarity 50, member B |
19410 |
0.19 |
| chr6_108910719_108911119 | 0.14 |
FOXO3 |
forkhead box O3 |
28850 |
0.24 |
| chr8_128004144_128004302 | 0.14 |
ENSG00000212451 |
. |
320456 |
0.01 |
| chr14_101034010_101035263 | 0.14 |
BEGAIN |
brain-enriched guanylate kinase-associated |
192 |
0.93 |
| chr1_198668823_198668976 | 0.14 |
RP11-553K8.5 |
|
32709 |
0.21 |
| chr21_37584630_37584785 | 0.14 |
ENSG00000265882 |
. |
1429 |
0.37 |
| chr5_96964556_96964707 | 0.14 |
CTD-2215E18.2 |
|
427047 |
0.01 |
| chr10_89628584_89628735 | 0.14 |
KLLN |
killin, p53-regulated DNA replication inhibitor |
5465 |
0.15 |
| chr1_45150633_45150798 | 0.14 |
C1orf228 |
chromosome 1 open reading frame 228 |
4791 |
0.13 |
| chr17_2117923_2118329 | 0.14 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
483 |
0.56 |
| chr7_76199840_76199991 | 0.14 |
AC004980.7 |
|
20980 |
0.16 |
| chr11_128116575_128116726 | 0.14 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
258639 |
0.02 |
| chr12_132469978_132470129 | 0.14 |
ENSG00000271905 |
. |
16267 |
0.15 |
| chr2_177392253_177392404 | 0.13 |
ENSG00000252027 |
. |
137076 |
0.04 |
| chrX_19902260_19902411 | 0.13 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
3242 |
0.33 |
| chr20_57623648_57623825 | 0.13 |
SLMO2 |
slowmo homolog 2 (Drosophila) |
5772 |
0.17 |
| chr1_38412223_38412952 | 0.13 |
INPP5B |
inositol polyphosphate-5-phosphatase, 75kDa |
136 |
0.94 |
| chr13_46979075_46979226 | 0.13 |
ENSG00000252385 |
. |
5439 |
0.21 |
| chr17_76743425_76743576 | 0.13 |
CYTH1 |
cytohesin 1 |
10528 |
0.2 |
| chr2_133998912_133999202 | 0.13 |
AC010890.1 |
|
23483 |
0.24 |
| chr5_167182705_167182856 | 0.13 |
TENM2 |
teneurin transmembrane protein 2 |
772 |
0.71 |
| chr21_46239564_46239715 | 0.13 |
SUMO3 |
small ubiquitin-like modifier 3 |
1595 |
0.28 |
| chr4_86788057_86788208 | 0.13 |
ARHGAP24 |
Rho GTPase activating protein 24 |
39086 |
0.17 |
| chr16_55116427_55116632 | 0.13 |
IRX5 |
iroquois homeobox 5 |
150545 |
0.04 |
| chr6_113012414_113012565 | 0.13 |
ENSG00000252215 |
. |
159105 |
0.04 |
| chr8_69905056_69905287 | 0.13 |
ENSG00000238808 |
. |
117638 |
0.07 |
| chr4_26289616_26289767 | 0.13 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
15462 |
0.3 |
| chr20_5574610_5574761 | 0.13 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
16987 |
0.24 |
| chr11_100586653_100586804 | 0.13 |
CTD-2383M3.1 |
|
28042 |
0.18 |
| chr8_94434922_94435073 | 0.13 |
LINC00535 |
long intergenic non-protein coding RNA 535 |
1889 |
0.41 |
| chr6_143897389_143897540 | 0.13 |
RP11-436I24.1 |
|
14296 |
0.19 |
| chrX_11229126_11229277 | 0.13 |
ARHGAP6 |
Rho GTPase activating protein 6 |
54894 |
0.14 |
| chr19_42083403_42083554 | 0.13 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
877 |
0.55 |
| chr7_92442804_92443186 | 0.12 |
CDK6 |
cyclin-dependent kinase 6 |
20236 |
0.2 |
| chr12_123201511_123201662 | 0.12 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
147 |
0.94 |
| chr18_4106677_4107316 | 0.12 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
44294 |
0.19 |
| chr15_101389305_101389898 | 0.12 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
28318 |
0.19 |
| chr4_52993404_52993555 | 0.12 |
SPATA18 |
spermatogenesis associated 18 |
75897 |
0.11 |
| chr1_164535920_164536071 | 0.12 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
3953 |
0.32 |
| chr11_122586485_122586636 | 0.12 |
ENSG00000239079 |
. |
10459 |
0.24 |
| chr7_15720774_15720925 | 0.12 |
MEOX2 |
mesenchyme homeobox 2 |
5588 |
0.26 |
| chr8_56793930_56794117 | 0.12 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
1629 |
0.31 |
| chr4_95635906_95636061 | 0.12 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
43136 |
0.21 |
| chr6_2478575_2478731 | 0.12 |
ENSG00000266252 |
. |
68784 |
0.13 |
| chr12_10281805_10282201 | 0.12 |
CLEC7A |
C-type lectin domain family 7, member A |
678 |
0.6 |
| chr3_148716623_148716961 | 0.12 |
GYG1 |
glycogenin 1 |
6634 |
0.21 |
| chr10_31271779_31271930 | 0.12 |
ZNF438 |
zinc finger protein 438 |
1501 |
0.54 |
| chr4_138309333_138309484 | 0.12 |
PCDH18 |
protocadherin 18 |
144157 |
0.05 |
| chr4_126108410_126108561 | 0.12 |
FAT4 |
FAT atypical cadherin 4 |
129069 |
0.06 |
| chr16_30997340_30997491 | 0.12 |
AC135048.1 |
Uncharacterized protein |
118 |
0.88 |
| chr5_53999711_53999862 | 0.12 |
ENSG00000221073 |
. |
148990 |
0.04 |
| chr6_134276070_134276235 | 0.12 |
TBPL1 |
TBP-like 1 |
1790 |
0.43 |
| chr9_94903241_94903667 | 0.12 |
ENSG00000238996 |
. |
4014 |
0.25 |
| chr19_31705372_31705523 | 0.11 |
AC020952.1 |
Uncharacterized protein |
65085 |
0.14 |
| chr9_92771713_92771864 | 0.11 |
ENSG00000263967 |
. |
14029 |
0.32 |
| chr1_84630171_84630563 | 0.11 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
3 |
0.99 |
| chrX_65233867_65234034 | 0.11 |
ENSG00000207939 |
. |
4762 |
0.23 |
| chr17_33703557_33703708 | 0.11 |
SLFN11 |
schlafen family member 11 |
2912 |
0.25 |
| chr10_32151742_32151893 | 0.11 |
ARHGAP12 |
Rho GTPase activating protein 12 |
8709 |
0.28 |
| chr1_150592184_150592335 | 0.11 |
ENSA |
endosulfine alpha |
9333 |
0.11 |
| chr12_19388273_19388424 | 0.11 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
1510 |
0.48 |
| chr7_133196546_133196697 | 0.11 |
EXOC4 |
exocyst complex component 4 |
64595 |
0.15 |
| chr5_35790158_35790403 | 0.11 |
SPEF2 |
sperm flagellar 2 |
11010 |
0.22 |
| chr15_49999586_49999737 | 0.11 |
RP11-353B9.1 |
|
55325 |
0.13 |
| chr1_192128120_192128535 | 0.11 |
RGS18 |
regulator of G-protein signaling 18 |
740 |
0.8 |
| chr1_72587992_72588143 | 0.11 |
NEGR1 |
neuronal growth regulator 1 |
21454 |
0.29 |
| chr5_92936152_92936303 | 0.11 |
ENSG00000237187 |
. |
14873 |
0.19 |
| chr7_134472487_134472744 | 0.11 |
CALD1 |
caldesmon 1 |
8186 |
0.3 |
| chr18_322790_322941 | 0.11 |
THOC1 |
THO complex 1 |
54838 |
0.14 |
| chr18_77526270_77526421 | 0.11 |
RP11-567M16.5 |
|
62694 |
0.12 |
| chr11_106239153_106239583 | 0.11 |
RP11-680E19.1 |
|
104326 |
0.08 |
| chr7_42183455_42183797 | 0.11 |
GLI3 |
GLI family zinc finger 3 |
83694 |
0.11 |
| chr4_54430281_54430491 | 0.11 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
6002 |
0.23 |
| chr12_123184919_123185107 | 0.11 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
2877 |
0.21 |
| chr4_102203498_102203727 | 0.11 |
ENSG00000221265 |
. |
47959 |
0.15 |
| chr10_109045223_109045524 | 0.11 |
SORCS1 |
sortilin-related VPS10 domain containing receptor 1 |
121081 |
0.07 |
| chr7_140048721_140049206 | 0.10 |
SLC37A3 |
solute carrier family 37, member 3 |
477 |
0.77 |
| chr1_169454193_169456202 | 0.10 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
28 |
0.98 |
| chr10_104912908_104913059 | 0.10 |
NT5C2 |
5'-nucleotidase, cytosolic II |
401 |
0.89 |
| chr6_113614101_113614252 | 0.10 |
ENSG00000222677 |
. |
222013 |
0.02 |
| chr1_200324732_200324883 | 0.10 |
ZNF281 |
zinc finger protein 281 |
54313 |
0.17 |
| chr8_68340215_68340366 | 0.10 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
84378 |
0.1 |
| chr17_33400140_33400291 | 0.10 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
9344 |
0.1 |
| chr1_170231057_170231208 | 0.10 |
ENSG00000263384 |
. |
108304 |
0.06 |
| chr1_156172023_156172174 | 0.10 |
SLC25A44 |
solute carrier family 25, member 44 |
8191 |
0.09 |
| chr7_87104246_87104923 | 0.10 |
ABCB4 |
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
197 |
0.96 |
| chr12_91743406_91743557 | 0.10 |
DCN |
decorin |
166581 |
0.04 |
| chr13_42422903_42423054 | 0.10 |
ENSG00000241406 |
. |
41165 |
0.17 |
| chr1_215179886_215180177 | 0.10 |
KCNK2 |
potassium channel, subfamily K, member 2 |
833 |
0.78 |
| chr17_69334209_69334649 | 0.10 |
ENSG00000222563 |
. |
27120 |
0.23 |
| chr7_17810311_17810462 | 0.10 |
SNX13 |
sorting nexin 13 |
169705 |
0.04 |
| chr3_11507449_11507639 | 0.10 |
ATG7 |
autophagy related 7 |
101411 |
0.07 |
| chr22_40046156_40046307 | 0.10 |
CACNA1I |
calcium channel, voltage-dependent, T type, alpha 1I subunit |
79473 |
0.08 |
| chr3_71151126_71151277 | 0.10 |
FOXP1 |
forkhead box P1 |
28543 |
0.26 |
| chr3_159628787_159629360 | 0.10 |
SCHIP1 |
schwannomin interacting protein 1 |
58345 |
0.11 |
| chr15_68433148_68433299 | 0.10 |
CALML4 |
calmodulin-like 4 |
59077 |
0.11 |
| chr5_66254399_66254664 | 0.10 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
167 |
0.97 |
| chr17_60882454_60882839 | 0.10 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
1355 |
0.53 |
| chr6_112008107_112008258 | 0.10 |
FYN |
FYN oncogene related to SRC, FGR, YES |
33083 |
0.16 |
| chr19_48751567_48751718 | 0.10 |
CARD8 |
caspase recruitment domain family, member 8 |
1283 |
0.32 |
| chr2_55519515_55519739 | 0.10 |
ENSG00000206964 |
. |
6473 |
0.14 |
| chr14_52819283_52819434 | 0.10 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
38245 |
0.19 |
| chr5_31197472_31197623 | 0.10 |
CDH6 |
cadherin 6, type 2, K-cadherin (fetal kidney) |
3690 |
0.35 |
| chr8_121352620_121352771 | 0.10 |
COL14A1 |
collagen, type XIV, alpha 1 |
4990 |
0.32 |
| chr18_11856994_11857516 | 0.10 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
184 |
0.63 |
| chr20_43344352_43344524 | 0.10 |
WISP2 |
WNT1 inducible signaling pathway protein 2 |
552 |
0.7 |
| chr8_23399467_23399639 | 0.10 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
12980 |
0.16 |
| chr9_79008529_79009235 | 0.10 |
RFK |
riboflavin kinase |
166 |
0.97 |
| chr13_52418432_52418583 | 0.10 |
CCDC70 |
coiled-coil domain containing 70 |
17610 |
0.19 |
| chr14_23116007_23116158 | 0.10 |
OR6J1 |
olfactory receptor, family 6, subfamily J, member 1 (gene/pseudogene) |
12366 |
0.14 |
| chr10_94516358_94516633 | 0.10 |
ENSG00000201412 |
. |
46130 |
0.14 |
| chr8_58660875_58661197 | 0.10 |
ENSG00000252057 |
. |
191892 |
0.03 |
| chr2_144704936_144705087 | 0.09 |
AC016910.1 |
|
10371 |
0.27 |
| chr9_71121412_71121563 | 0.09 |
TMEM252 |
transmembrane protein 252 |
34296 |
0.18 |
| chr5_139203558_139203709 | 0.09 |
CTB-35F21.4 |
|
24242 |
0.17 |
| chr1_57092584_57092735 | 0.09 |
PRKAA2 |
protein kinase, AMP-activated, alpha 2 catalytic subunit |
18336 |
0.24 |
| chr6_20497460_20497742 | 0.09 |
ENSG00000201683 |
. |
2805 |
0.26 |
| chr19_18428816_18429000 | 0.09 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
1839 |
0.19 |
| chr12_9946809_9947109 | 0.09 |
KLRF1 |
killer cell lectin-like receptor subfamily F, member 1 |
33118 |
0.11 |
| chrX_10723894_10724086 | 0.09 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
78211 |
0.12 |
| chr2_71858264_71858415 | 0.09 |
DYSF |
dysferlin |
164507 |
0.04 |
| chr13_42143168_42143319 | 0.09 |
ENSG00000264190 |
. |
712 |
0.76 |
| chr3_32433354_32434295 | 0.09 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
293 |
0.93 |
| chr3_73862389_73862540 | 0.09 |
PDZRN3 |
PDZ domain containing ring finger 3 |
188373 |
0.03 |
| chr1_120188688_120188839 | 0.09 |
ZNF697 |
zinc finger protein 697 |
1633 |
0.39 |
| chr11_28127734_28127940 | 0.09 |
METTL15 |
methyltransferase like 15 |
1958 |
0.31 |
| chr10_30002863_30003014 | 0.09 |
ENSG00000222092 |
. |
2094 |
0.38 |
| chr12_82336788_82336939 | 0.09 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
183531 |
0.03 |
| chr22_39149918_39150069 | 0.09 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
787 |
0.4 |
| chr10_73486217_73486453 | 0.09 |
C10orf105 |
chromosome 10 open reading frame 105 |
6757 |
0.23 |
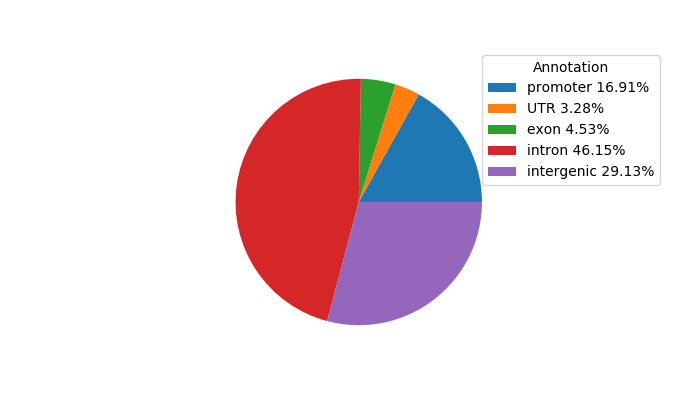
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0060557 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.0 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:1901889 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |