Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
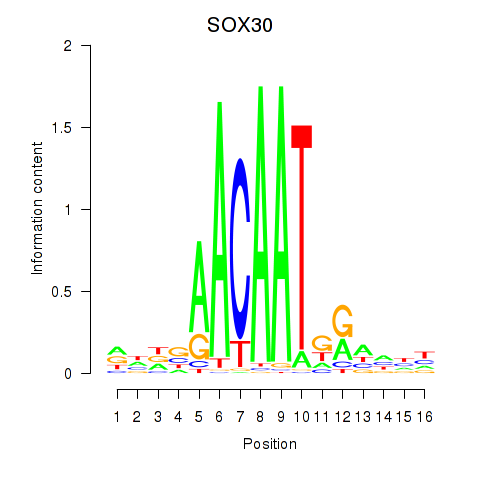
Results for SOX30
Z-value: 0.86
Transcription factors associated with SOX30
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX30
|
ENSG00000039600.6 | SRY-box transcription factor 30 |
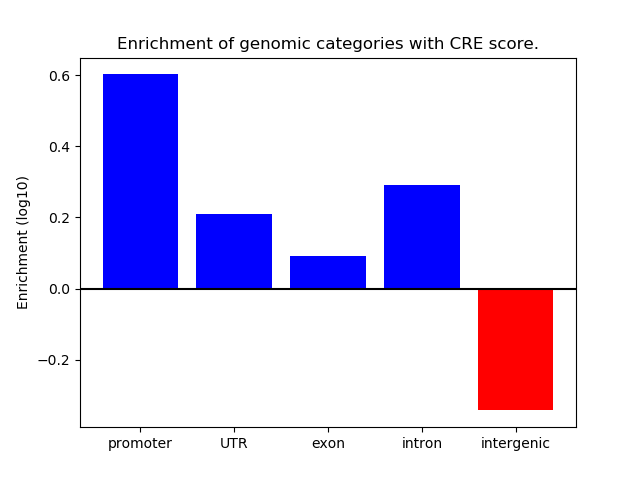
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_157078570_157078774 | SOX30 | 700 | 0.630058 | -0.73 | 2.4e-02 | Click! |
| chr5_157078809_157079318 | SOX30 | 309 | 0.871837 | -0.65 | 5.6e-02 | Click! |
| chr5_157097546_157097697 | SOX30 | 867 | 0.378634 | 0.55 | 1.3e-01 | Click! |
| chr5_157097802_157098814 | SOX30 | 180 | 0.587499 | -0.51 | 1.6e-01 | Click! |
| chr5_157079325_157079476 | SOX30 | 28 | 0.971191 | -0.26 | 5.0e-01 | Click! |
Activity of the SOX30 motif across conditions
Conditions sorted by the z-value of the SOX30 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
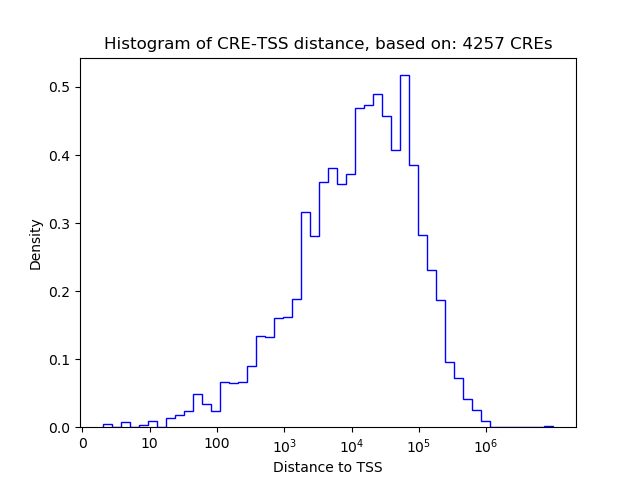
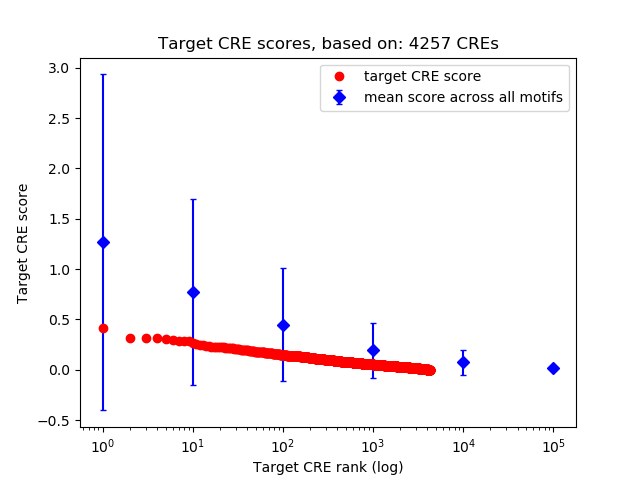
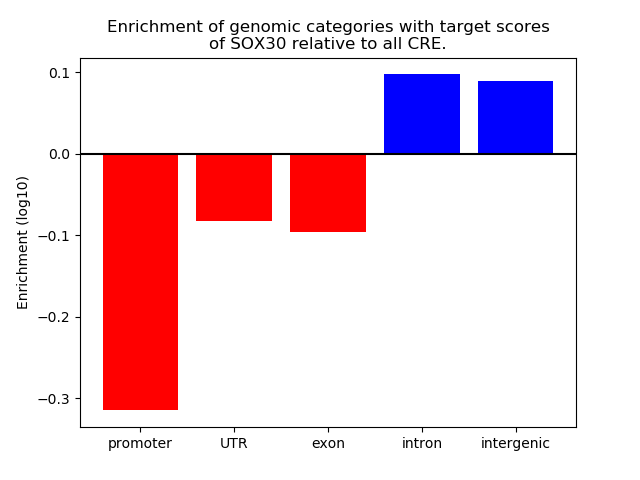
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_91570174_91570436 | 0.41 |
DCN |
decorin |
2024 |
0.44 |
| chr9_73095232_73095769 | 0.32 |
KLF9 |
Kruppel-like factor 9 |
65960 |
0.12 |
| chr7_93465582_93465846 | 0.32 |
TFPI2 |
tissue factor pathway inhibitor 2 |
53768 |
0.12 |
| chr14_73881950_73882101 | 0.32 |
NUMB |
numb homolog (Drosophila) |
5246 |
0.21 |
| chr19_34359259_34359976 | 0.30 |
ENSG00000240626 |
. |
58913 |
0.13 |
| chr10_62547859_62548079 | 0.30 |
CDK1 |
cyclin-dependent kinase 1 |
8058 |
0.28 |
| chr18_56528219_56528370 | 0.29 |
ZNF532 |
zinc finger protein 532 |
1538 |
0.39 |
| chr16_3501012_3501163 | 0.28 |
NAA60 |
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
5644 |
0.13 |
| chr11_82608630_82608781 | 0.28 |
C11orf82 |
chromosome 11 open reading frame 82 |
2312 |
0.28 |
| chr7_98719396_98719594 | 0.26 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
22147 |
0.2 |
| chr15_92404035_92404308 | 0.25 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
6820 |
0.27 |
| chr7_81679403_81679554 | 0.25 |
MIR1255B1 |
microRNA 1255b-1 |
40970 |
0.18 |
| chr18_46460407_46461027 | 0.24 |
SMAD7 |
SMAD family member 7 |
14158 |
0.24 |
| chr7_15723868_15724269 | 0.24 |
MEOX2 |
mesenchyme homeobox 2 |
2369 |
0.37 |
| chr13_86099472_86099623 | 0.23 |
ENSG00000207012 |
. |
59441 |
0.17 |
| chr4_75409052_75409203 | 0.23 |
RP11-727M10.2 |
|
66845 |
0.1 |
| chr9_18476160_18476529 | 0.22 |
ADAMTSL1 |
ADAMTS-like 1 |
2113 |
0.45 |
| chr2_190075884_190076035 | 0.22 |
COL5A2 |
collagen, type V, alpha 2 |
31354 |
0.2 |
| chr3_187984581_187985052 | 0.22 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
27164 |
0.24 |
| chr5_180682825_180682976 | 0.22 |
AC008443.1 |
CDNA: FLJ23158 fis, clone LNG09623; Uncharacterized protein |
180 |
0.86 |
| chr4_82326915_82327066 | 0.22 |
RASGEF1B |
RasGEF domain family, member 1B |
65400 |
0.13 |
| chr13_24807615_24808021 | 0.22 |
SPATA13 |
spermatogenesis associated 13 |
18014 |
0.17 |
| chr8_40602961_40603112 | 0.22 |
ZMAT4 |
zinc finger, matrin-type 4 |
143028 |
0.05 |
| chr2_227590884_227591406 | 0.22 |
ENSG00000263363 |
. |
67636 |
0.11 |
| chr6_126124183_126124375 | 0.22 |
NCOA7 |
nuclear receptor coactivator 7 |
7002 |
0.19 |
| chr1_153582582_153582782 | 0.21 |
S100A16 |
S100 calcium binding protein A16 |
862 |
0.35 |
| chr4_36257830_36257981 | 0.21 |
RP11-431M7.3 |
|
235 |
0.94 |
| chr11_46402058_46403020 | 0.21 |
MDK |
midkine (neurite growth-promoting factor 2) |
52 |
0.96 |
| chrX_39949292_39950247 | 0.21 |
BCOR |
BCL6 corepressor |
6887 |
0.33 |
| chr3_154959930_154960081 | 0.21 |
MME-AS1 |
MME antisense RNA 1 |
58931 |
0.15 |
| chr12_110993281_110993585 | 0.20 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
27692 |
0.12 |
| chr2_50371364_50371541 | 0.20 |
NRXN1 |
neurexin 1 |
125041 |
0.06 |
| chr4_38705158_38705309 | 0.20 |
RP11-617D20.1 |
|
38729 |
0.12 |
| chr7_17080807_17081087 | 0.20 |
AGR3 |
anterior gradient 3 |
159336 |
0.04 |
| chr4_118249233_118249384 | 0.20 |
TRAM1L1 |
translocation associated membrane protein 1-like 1 |
242572 |
0.02 |
| chr8_97145200_97145762 | 0.20 |
GDF6 |
growth differentiation factor 6 |
27539 |
0.21 |
| chr10_4358088_4358256 | 0.20 |
ENSG00000207124 |
. |
198972 |
0.03 |
| chr3_113460155_113460306 | 0.20 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
1796 |
0.34 |
| chr7_116164164_116164509 | 0.20 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
503 |
0.76 |
| chr3_56929337_56929488 | 0.20 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
21087 |
0.22 |
| chr1_161477171_161477322 | 0.19 |
FCGR2A |
Fc fragment of IgG, low affinity IIa, receptor (CD32) |
2026 |
0.25 |
| chr18_61496009_61496309 | 0.19 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
42767 |
0.13 |
| chr19_45959494_45959730 | 0.19 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
5629 |
0.12 |
| chr2_237884917_237885068 | 0.19 |
ENSG00000202341 |
. |
38956 |
0.18 |
| chr9_128521294_128521792 | 0.19 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
11065 |
0.27 |
| chr4_54564515_54564666 | 0.19 |
RP11-317M11.1 |
|
2522 |
0.3 |
| chr8_25442204_25442403 | 0.18 |
CDCA2 |
cell division cycle associated 2 |
125588 |
0.06 |
| chr22_30588637_30588788 | 0.18 |
RP3-438O4.4 |
|
14386 |
0.14 |
| chr6_25360210_25360380 | 0.18 |
ENSG00000207490 |
. |
23532 |
0.16 |
| chr12_69809879_69810030 | 0.18 |
ENSG00000266185 |
. |
15271 |
0.18 |
| chr2_238070262_238070413 | 0.18 |
COPS8 |
COP9 signalosome subunit 8 |
75751 |
0.09 |
| chr2_189842191_189842342 | 0.18 |
ENSG00000221502 |
. |
552 |
0.78 |
| chr2_113537340_113537491 | 0.18 |
IL1A |
interleukin 1, alpha |
4752 |
0.19 |
| chr6_7692698_7692863 | 0.18 |
BMP6 |
bone morphogenetic protein 6 |
34250 |
0.21 |
| chr2_226029710_226029861 | 0.18 |
DOCK10 |
dedicator of cytokinesis 10 |
122626 |
0.06 |
| chr18_38705827_38705978 | 0.18 |
ENSG00000238333 |
. |
653117 |
0.0 |
| chr12_18318254_18318405 | 0.18 |
RERGL |
RERG/RAS-like |
75202 |
0.12 |
| chr1_155469525_155469676 | 0.18 |
ENSG00000235919 |
. |
62264 |
0.07 |
| chr12_65550856_65551007 | 0.17 |
LEMD3 |
LEM domain containing 3 |
12420 |
0.21 |
| chr11_88067311_88067773 | 0.17 |
CTSC |
cathepsin C |
3359 |
0.37 |
| chr14_41685035_41685186 | 0.17 |
ENSG00000221695 |
. |
133387 |
0.06 |
| chr10_112627670_112627821 | 0.17 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
3820 |
0.17 |
| chr10_123909165_123909405 | 0.17 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
13656 |
0.27 |
| chr3_157822422_157822707 | 0.17 |
SHOX2 |
short stature homeobox 2 |
519 |
0.7 |
| chr12_119617823_119618008 | 0.17 |
HSPB8 |
heat shock 22kDa protein 8 |
520 |
0.73 |
| chr12_14569534_14569685 | 0.17 |
ATF7IP |
activating transcription factor 7 interacting protein |
238 |
0.95 |
| chr6_64062151_64062302 | 0.17 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
32344 |
0.24 |
| chr2_175743756_175743907 | 0.17 |
CHN1 |
chimerin 1 |
31352 |
0.19 |
| chr11_111047528_111047679 | 0.17 |
C11orf53 |
chromosome 11 open reading frame 53 |
79104 |
0.09 |
| chr3_29991791_29991942 | 0.17 |
RBMS3-AS1 |
RBMS3 antisense RNA 1 |
16219 |
0.29 |
| chr7_133055658_133055809 | 0.17 |
ENSG00000238844 |
. |
108248 |
0.07 |
| chr5_126348889_126349040 | 0.17 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
17536 |
0.24 |
| chr10_29229083_29229234 | 0.17 |
ENSG00000199402 |
. |
65722 |
0.13 |
| chr9_13568681_13569117 | 0.17 |
MPDZ |
multiple PDZ domain protein |
289310 |
0.01 |
| chrX_50556610_50557434 | 0.17 |
SHROOM4 |
shroom family member 4 |
22 |
0.99 |
| chr8_79468522_79468695 | 0.16 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
34858 |
0.19 |
| chr3_10315902_10316053 | 0.16 |
TATDN2 |
TatD DNase domain containing 2 |
3373 |
0.15 |
| chr9_133713242_133713393 | 0.16 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
2864 |
0.3 |
| chr10_30346002_30346153 | 0.16 |
KIAA1462 |
KIAA1462 |
2376 |
0.44 |
| chr17_67650795_67650946 | 0.16 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
152300 |
0.04 |
| chr13_36931940_36932091 | 0.16 |
ENSG00000266047 |
. |
2087 |
0.25 |
| chr7_120664705_120664856 | 0.16 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
35104 |
0.16 |
| chr3_56810315_56810466 | 0.16 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
705 |
0.79 |
| chr10_114714851_114715858 | 0.16 |
RP11-57H14.2 |
|
3720 |
0.26 |
| chr2_73021663_73021814 | 0.16 |
EXOC6B |
exocyst complex component 6B |
31432 |
0.17 |
| chr7_92467890_92468041 | 0.16 |
CDK6 |
cyclin-dependent kinase 6 |
2057 |
0.31 |
| chr16_3911841_3912078 | 0.16 |
CREBBP |
CREB binding protein |
18162 |
0.21 |
| chr5_141218824_141218975 | 0.16 |
PCDH1 |
protocadherin 1 |
30255 |
0.16 |
| chr7_19136246_19136397 | 0.15 |
AC003986.6 |
|
15776 |
0.16 |
| chr2_162019757_162019908 | 0.15 |
TANK |
TRAF family member-associated NFKB activator |
2595 |
0.29 |
| chr15_52944945_52945096 | 0.15 |
FAM214A |
family with sequence similarity 214, member A |
773 |
0.64 |
| chr3_74475331_74475482 | 0.15 |
CNTN3 |
contactin 3 (plasmacytoma associated) |
94885 |
0.09 |
| chr20_60594176_60594409 | 0.15 |
TAF4 |
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
11675 |
0.2 |
| chr2_19995754_19995974 | 0.15 |
TTC32 |
tetratricopeptide repeat domain 32 |
105538 |
0.07 |
| chr6_70501419_70501570 | 0.15 |
LMBRD1 |
LMBR1 domain containing 1 |
937 |
0.71 |
| chr16_79732693_79732844 | 0.15 |
ENSG00000221330 |
. |
29134 |
0.25 |
| chr10_44789701_44789852 | 0.15 |
RP11-20J15.3 |
|
619 |
0.82 |
| chrX_115371447_115371702 | 0.15 |
AGTR2 |
angiotensin II receptor, type 2 |
69599 |
0.13 |
| chr21_34602307_34603720 | 0.15 |
IFNAR2 |
interferon (alpha, beta and omega) receptor 2 |
299 |
0.87 |
| chr6_1613704_1614304 | 0.15 |
FOXC1 |
forkhead box C1 |
3323 |
0.37 |
| chr5_40487521_40487672 | 0.15 |
ENSG00000265615 |
. |
167176 |
0.03 |
| chr12_54698226_54698575 | 0.15 |
COPZ1 |
coatomer protein complex, subunit zeta 1 |
3414 |
0.1 |
| chr1_36183617_36184771 | 0.15 |
C1orf216 |
chromosome 1 open reading frame 216 |
879 |
0.53 |
| chr1_167132672_167132823 | 0.15 |
RP11-277B15.2 |
|
31970 |
0.14 |
| chr6_45463009_45463160 | 0.14 |
RUNX2 |
runt-related transcription factor 2 |
72862 |
0.11 |
| chr1_54232914_54233120 | 0.14 |
ENSG00000201003 |
. |
3809 |
0.22 |
| chr5_134844161_134844312 | 0.14 |
NEUROG1 |
neurogenin 1 |
27403 |
0.14 |
| chr12_19375788_19375939 | 0.14 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
13995 |
0.22 |
| chr1_210057563_210057765 | 0.14 |
DIEXF |
digestive organ expansion factor homolog (zebrafish) |
45514 |
0.13 |
| chr9_113940701_113941000 | 0.14 |
ENSG00000212409 |
. |
81157 |
0.1 |
| chr3_70854632_70854783 | 0.14 |
ENSG00000206939 |
. |
2794 |
0.39 |
| chr7_91195255_91195601 | 0.14 |
FZD1 |
frizzled family receptor 1 |
301645 |
0.01 |
| chr10_33239282_33239433 | 0.14 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
5234 |
0.31 |
| chr4_77506894_77507309 | 0.14 |
ENSG00000265314 |
. |
10397 |
0.17 |
| chr3_173460576_173460750 | 0.14 |
NLGN1 |
neuroligin 1 |
138274 |
0.05 |
| chr17_14642934_14643085 | 0.14 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
438609 |
0.01 |
| chr1_231648231_231648382 | 0.14 |
RP11-295G20.2 |
|
15695 |
0.19 |
| chr10_18448210_18448361 | 0.14 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
18176 |
0.26 |
| chr12_56959274_56959425 | 0.14 |
ENSG00000201579 |
. |
22953 |
0.12 |
| chr15_57509304_57509455 | 0.14 |
TCF12 |
transcription factor 12 |
2249 |
0.41 |
| chr3_158486598_158486991 | 0.14 |
RP11-379F4.8 |
|
13542 |
0.14 |
| chr7_39504844_39505021 | 0.14 |
POU6F2-AS1 |
POU6F2 antisense RNA 1 |
58987 |
0.12 |
| chr2_37814036_37814187 | 0.14 |
AC006369.2 |
|
13168 |
0.25 |
| chr18_10101229_10101442 | 0.14 |
ENSG00000263630 |
. |
96141 |
0.08 |
| chr11_17369748_17369899 | 0.14 |
NCR3LG1 |
natural killer cell cytotoxicity receptor 3 ligand 1 |
3450 |
0.21 |
| chr6_85482826_85483217 | 0.14 |
TBX18 |
T-box 18 |
8784 |
0.3 |
| chr1_170641859_170642010 | 0.14 |
PRRX1 |
paired related homeobox 1 |
8856 |
0.29 |
| chr15_40111813_40111964 | 0.14 |
RP11-37C7.2 |
|
4258 |
0.19 |
| chr12_11916833_11916984 | 0.14 |
ETV6 |
ets variant 6 |
11473 |
0.29 |
| chr8_13347325_13347476 | 0.14 |
DLC1 |
deleted in liver cancer 1 |
9350 |
0.24 |
| chr1_35654542_35654817 | 0.14 |
SFPQ |
splicing factor proline/glutamine-rich |
4070 |
0.21 |
| chr5_56600937_56601088 | 0.14 |
CTD-2023N9.1 |
|
89875 |
0.08 |
| chr9_71802023_71802174 | 0.14 |
TJP2 |
tight junction protein 2 |
12888 |
0.25 |
| chr19_58488964_58489115 | 0.14 |
C19orf18 |
chromosome 19 open reading frame 18 |
3137 |
0.15 |
| chr2_45004607_45004758 | 0.14 |
ENSG00000252896 |
. |
4587 |
0.34 |
| chr9_75192112_75192273 | 0.14 |
TMC1 |
transmembrane channel-like 1 |
37424 |
0.19 |
| chr1_235115156_235115307 | 0.14 |
ENSG00000239690 |
. |
75298 |
0.1 |
| chr9_26170624_26170775 | 0.14 |
ENSG00000266429 |
. |
443424 |
0.01 |
| chr6_129203821_129204789 | 0.14 |
LAMA2 |
laminin, alpha 2 |
37 |
0.99 |
| chr17_8263383_8263534 | 0.14 |
AC135178.1 |
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
80 |
0.93 |
| chr5_142709588_142709739 | 0.14 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
70754 |
0.12 |
| chr6_130391698_130391967 | 0.13 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
50130 |
0.15 |
| chr3_124510640_124510940 | 0.13 |
ITGB5-AS1 |
ITGB5 antisense RNA 1 |
10788 |
0.18 |
| chr2_148122279_148122430 | 0.13 |
ENSG00000238860 |
. |
40815 |
0.16 |
| chr3_188311777_188312056 | 0.13 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
15093 |
0.2 |
| chr3_164178912_164179063 | 0.13 |
ENSG00000221755 |
. |
119858 |
0.07 |
| chr15_75870175_75871107 | 0.13 |
PTPN9 |
protein tyrosine phosphatase, non-receptor type 9 |
989 |
0.47 |
| chr18_9918630_9918781 | 0.13 |
VAPA |
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa |
4646 |
0.2 |
| chr4_34171764_34171915 | 0.13 |
ENSG00000263814 |
. |
216620 |
0.02 |
| chr10_36066984_36067135 | 0.13 |
FZD8 |
frizzled family receptor 8 |
136697 |
0.05 |
| chr3_120215962_120216316 | 0.13 |
FSTL1 |
follistatin-like 1 |
46039 |
0.16 |
| chr6_160106825_160107100 | 0.13 |
SOD2 |
superoxide dismutase 2, mitochondrial |
7298 |
0.18 |
| chr1_225923826_225924038 | 0.13 |
RP11-145A3.2 |
|
44 |
0.97 |
| chr17_73386962_73387166 | 0.13 |
GRB2 |
growth factor receptor-bound protein 2 |
2673 |
0.13 |
| chr1_36549458_36550287 | 0.13 |
TEKT2 |
tektin 2 (testicular) |
196 |
0.92 |
| chr4_6909699_6909850 | 0.13 |
TBC1D14 |
TBC1 domain family, member 14 |
1195 |
0.49 |
| chr2_225734023_225734174 | 0.13 |
DOCK10 |
dedicator of cytokinesis 10 |
52729 |
0.18 |
| chr7_152157267_152157418 | 0.13 |
ENSG00000221454 |
. |
21036 |
0.18 |
| chr6_83077930_83078081 | 0.13 |
TPBG |
trophoblast glycoprotein |
4044 |
0.35 |
| chr15_94841194_94841722 | 0.13 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
28 |
0.99 |
| chr4_183413823_183413976 | 0.13 |
TENM3 |
teneurin transmembrane protein 3 |
43747 |
0.17 |
| chr8_77690391_77690542 | 0.13 |
ZFHX4 |
zinc finger homeobox 4 |
74186 |
0.11 |
| chr2_1711742_1712354 | 0.13 |
PXDN |
peroxidasin homolog (Drosophila) |
15706 |
0.26 |
| chr4_26022444_26022665 | 0.13 |
SMIM20 |
small integral membrane protein 20 |
106623 |
0.07 |
| chr7_35936307_35936458 | 0.13 |
SEPT7 |
septin 7 |
16856 |
0.2 |
| chr9_132246124_132246361 | 0.13 |
ENSG00000264298 |
. |
5407 |
0.24 |
| chr6_169426238_169426389 | 0.13 |
XXyac-YX65C7_A.2 |
|
187036 |
0.03 |
| chr12_118626563_118627015 | 0.13 |
TAOK3 |
TAO kinase 3 |
1538 |
0.42 |
| chr2_134484282_134484433 | 0.13 |
ENSG00000200708 |
. |
130695 |
0.05 |
| chr5_77935751_77935902 | 0.13 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
8822 |
0.32 |
| chr8_42054040_42054280 | 0.13 |
PLAT |
plasminogen activator, tissue |
944 |
0.52 |
| chr9_81972295_81972446 | 0.13 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
214318 |
0.03 |
| chr3_168866326_168866618 | 0.13 |
MECOM |
MDS1 and EVI1 complex locus |
950 |
0.73 |
| chr21_28339413_28339697 | 0.13 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
723 |
0.73 |
| chr19_10678474_10679697 | 0.13 |
CDKN2D |
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
569 |
0.54 |
| chr16_27005073_27005435 | 0.13 |
RP11-673P17.2 |
|
73925 |
0.12 |
| chr12_81616950_81617101 | 0.13 |
ENSG00000265227 |
. |
64858 |
0.11 |
| chr17_28687356_28687507 | 0.13 |
ENSG00000201255 |
. |
50 |
0.96 |
| chr3_149371362_149371801 | 0.13 |
WWTR1-AS1 |
WWTR1 antisense RNA 1 |
3226 |
0.2 |
| chr9_90427762_90427913 | 0.13 |
XXyac-YM21GA2.6 |
|
24303 |
0.14 |
| chr13_41540297_41540448 | 0.13 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
16046 |
0.18 |
| chr14_47696792_47696943 | 0.13 |
MDGA2 |
MAM domain containing glycosylphosphatidylinositol anchor 2 |
95826 |
0.09 |
| chr16_19769806_19769957 | 0.12 |
CTD-2380F24.1 |
|
7485 |
0.18 |
| chr13_108536105_108536256 | 0.12 |
FAM155A |
family with sequence similarity 155, member A |
17097 |
0.25 |
| chr18_66506011_66506162 | 0.12 |
CCDC102B |
coiled-coil domain containing 102B |
2085 |
0.36 |
| chr6_39398889_39399040 | 0.12 |
KIF6 |
kinesin family member 6 |
125 |
0.98 |
| chr15_38556619_38556770 | 0.12 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
11312 |
0.31 |
| chr2_62705563_62705714 | 0.12 |
ENSG00000241625 |
. |
12675 |
0.23 |
| chr5_57793978_57794129 | 0.12 |
GAPT |
GRB2-binding adaptor protein, transmembrane |
3986 |
0.24 |
| chr4_81195528_81195918 | 0.12 |
FGF5 |
fibroblast growth factor 5 |
7930 |
0.26 |
| chr14_27311618_27311769 | 0.12 |
NOVA1-AS1 |
NOVA1 antisense RNA 1 (head to head) |
60319 |
0.13 |
| chr5_39275351_39275502 | 0.12 |
FYB |
FYN binding protein |
796 |
0.75 |
| chr7_111720888_111721301 | 0.12 |
DOCK4 |
dedicator of cytokinesis 4 |
76908 |
0.11 |
| chr12_116510107_116510258 | 0.12 |
ENSG00000200665 |
. |
10193 |
0.25 |
| chr3_87000039_87000329 | 0.12 |
VGLL3 |
vestigial like 3 (Drosophila) |
39668 |
0.23 |
| chr10_8343377_8343528 | 0.12 |
GATA3 |
GATA binding protein 3 |
246683 |
0.02 |
| chr1_247714015_247714166 | 0.12 |
GCSAML |
germinal center-associated, signaling and motility-like |
1641 |
0.26 |
| chr4_169769420_169769799 | 0.12 |
RP11-635L1.3 |
|
15521 |
0.19 |
| chr8_108344303_108344567 | 0.12 |
ANGPT1 |
angiopoietin 1 |
4315 |
0.34 |
| chr7_132339761_132340479 | 0.12 |
AC009365.3 |
|
6567 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.0 | 0.0 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.0 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0090030 | regulation of steroid hormone biosynthetic process(GO:0090030) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |