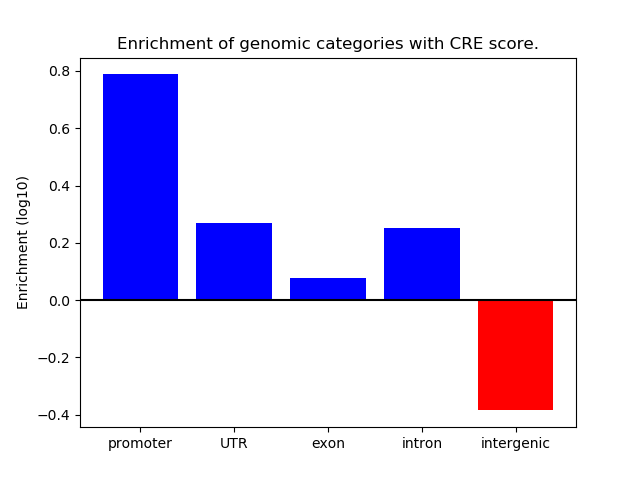
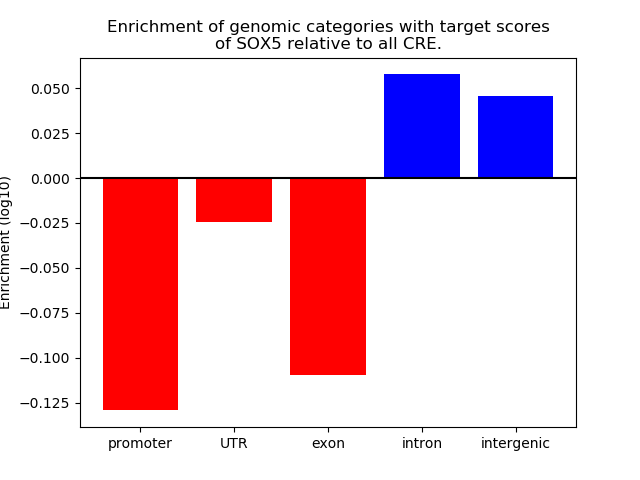
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
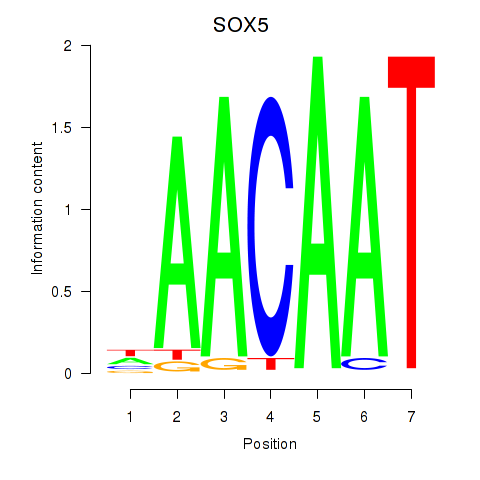
Results for SOX5
Z-value: 1.18
Transcription factors associated with SOX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX5
|
ENSG00000134532.11 | SRY-box transcription factor 5 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_24102871_24103025 | SOX5 | 311 | 0.948913 | 0.70 | 3.7e-02 | Click! |
| chr12_24103066_24103383 | SOX5 | 587 | 0.864093 | 0.65 | 5.8e-02 | Click! |
| chr12_24102324_24102475 | SOX5 | 182 | 0.975163 | 0.65 | 5.9e-02 | Click! |
| chr12_24199012_24199210 | SOX5 | 95145 | 0.092801 | 0.62 | 7.3e-02 | Click! |
| chr12_24048929_24049080 | SOX5 | 43 | 0.988942 | 0.52 | 1.5e-01 | Click! |
Activity of the SOX5 motif across conditions
Conditions sorted by the z-value of the SOX5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_114714851_114715858 | 0.43 |
RP11-57H14.2 |
|
3720 |
0.26 |
| chr11_131780761_131781452 | 0.36 |
NTM |
neurotrimin |
209 |
0.95 |
| chr4_55095719_55096018 | 0.35 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
142 |
0.98 |
| chr6_27584810_27585278 | 0.33 |
ENSG00000238648 |
. |
14144 |
0.19 |
| chr8_19148668_19149118 | 0.33 |
SH2D4A |
SH2 domain containing 4A |
22235 |
0.28 |
| chr15_49716260_49716444 | 0.31 |
FGF7 |
fibroblast growth factor 7 |
895 |
0.66 |
| chr2_189839524_189839773 | 0.31 |
COL3A1 |
collagen, type III, alpha 1 |
549 |
0.79 |
| chr3_87036535_87037613 | 0.29 |
VGLL3 |
vestigial like 3 (Drosophila) |
2778 |
0.42 |
| chr7_93552303_93552503 | 0.28 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
1392 |
0.38 |
| chr7_19155713_19156221 | 0.28 |
TWIST1 |
twist family bHLH transcription factor 1 |
1328 |
0.36 |
| chr18_31157392_31158104 | 0.27 |
ASXL3 |
additional sex combs like 3 (Drosophila) |
848 |
0.76 |
| chr8_142289533_142290316 | 0.27 |
RP11-10J21.3 |
Uncharacterized protein |
25260 |
0.12 |
| chr2_189841718_189841869 | 0.26 |
ENSG00000221502 |
. |
1025 |
0.55 |
| chr1_178064247_178064419 | 0.25 |
RASAL2 |
RAS protein activator like 2 |
1057 |
0.69 |
| chr2_9815742_9816384 | 0.23 |
YWHAQ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
44920 |
0.13 |
| chr11_121964934_121965228 | 0.23 |
ENSG00000207971 |
. |
5471 |
0.18 |
| chr9_133656128_133656348 | 0.23 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
54215 |
0.13 |
| chr1_172110530_172111247 | 0.23 |
ENSG00000207949 |
. |
2841 |
0.22 |
| chr7_127307743_127307970 | 0.22 |
SND1 |
staphylococcal nuclease and tudor domain containing 1 |
15622 |
0.17 |
| chr11_121800761_121801203 | 0.22 |
ENSG00000252556 |
. |
74081 |
0.11 |
| chr5_134825154_134825603 | 0.22 |
TIFAB |
TRAF-interacting protein with forkhead-associated domain, family member B |
37289 |
0.11 |
| chr10_69831641_69832005 | 0.21 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
2071 |
0.33 |
| chr11_9548903_9549060 | 0.21 |
ZNF143 |
zinc finger protein 143 |
14937 |
0.16 |
| chr6_85482826_85483217 | 0.21 |
TBX18 |
T-box 18 |
8784 |
0.3 |
| chr4_159093168_159093728 | 0.21 |
RP11-597D13.9 |
|
281 |
0.64 |
| chr9_22008020_22008450 | 0.20 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
717 |
0.61 |
| chr3_16217443_16217716 | 0.20 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
1363 |
0.53 |
| chr10_6763782_6764322 | 0.20 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr9_6218213_6218406 | 0.20 |
IL33 |
interleukin 33 |
2500 |
0.39 |
| chr13_36047635_36048256 | 0.20 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
2887 |
0.26 |
| chrX_78003533_78003765 | 0.20 |
LPAR4 |
lysophosphatidic acid receptor 4 |
421 |
0.91 |
| chr4_186696107_186696537 | 0.20 |
SORBS2 |
sorbin and SH3 domain containing 2 |
108 |
0.98 |
| chr7_90012947_90013805 | 0.20 |
CLDN12 |
claudin 12 |
341 |
0.9 |
| chr4_18595427_18595619 | 0.19 |
LCORL |
ligand dependent nuclear receptor corepressor-like |
572024 |
0.0 |
| chr8_93115764_93116215 | 0.19 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
475 |
0.9 |
| chr1_101313202_101313363 | 0.19 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
46938 |
0.12 |
| chr2_54195795_54196058 | 0.18 |
ACYP2 |
acylphosphatase 2, muscle type |
2049 |
0.28 |
| chrX_139847648_139847972 | 0.18 |
CDR1 |
cerebellar degeneration-related protein 1, 34kDa |
18913 |
0.23 |
| chr6_53934757_53935103 | 0.18 |
MLIP-AS1 |
MLIP antisense RNA 1 |
9594 |
0.22 |
| chr2_152214605_152215087 | 0.18 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
740 |
0.63 |
| chr8_72273557_72273741 | 0.18 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
446 |
0.9 |
| chr3_120215962_120216316 | 0.18 |
FSTL1 |
follistatin-like 1 |
46039 |
0.16 |
| chr9_133712337_133712924 | 0.18 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
2177 |
0.35 |
| chr4_86397582_86397737 | 0.18 |
ARHGAP24 |
Rho GTPase activating protein 24 |
1268 |
0.64 |
| chr12_88971091_88971358 | 0.17 |
KITLG |
KIT ligand |
3014 |
0.3 |
| chr7_30636157_30636308 | 0.17 |
ENSG00000196295 |
. |
1807 |
0.29 |
| chr1_94882933_94883167 | 0.17 |
ABCD3 |
ATP-binding cassette, sub-family D (ALD), member 3 |
883 |
0.72 |
| chr18_12076288_12076458 | 0.17 |
ANKRD62 |
ankyrin repeat domain 62 |
17479 |
0.16 |
| chr8_107738861_107739281 | 0.17 |
OXR1 |
oxidation resistance 1 |
658 |
0.71 |
| chr1_56136277_56136451 | 0.17 |
ENSG00000272051 |
. |
185719 |
0.03 |
| chr6_3749707_3750198 | 0.17 |
RP11-420L9.5 |
|
1393 |
0.41 |
| chr12_89619824_89620001 | 0.17 |
ENSG00000238302 |
. |
56150 |
0.16 |
| chr12_78336681_78336916 | 0.17 |
NAV3 |
neuron navigator 3 |
23258 |
0.28 |
| chr9_12776574_12776837 | 0.17 |
LURAP1L |
leucine rich adaptor protein 1-like |
1685 |
0.38 |
| chr12_117036919_117037510 | 0.17 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
23558 |
0.24 |
| chr6_150176715_150177267 | 0.17 |
RP11-350J20.12 |
|
3393 |
0.15 |
| chr4_102277856_102278206 | 0.17 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
8596 |
0.2 |
| chr2_175711229_175711408 | 0.17 |
CHN1 |
chimerin 1 |
175 |
0.97 |
| chr15_61326560_61326811 | 0.16 |
RP11-39M21.1 |
|
146763 |
0.04 |
| chr1_179279550_179279920 | 0.16 |
SOAT1 |
sterol O-acyltransferase 1 |
16280 |
0.19 |
| chr6_135679919_135680436 | 0.16 |
AHI1 |
Abelson helper integration site 1 |
52469 |
0.13 |
| chr6_53928343_53928541 | 0.16 |
MLIP-AS1 |
MLIP antisense RNA 1 |
16050 |
0.2 |
| chr6_12491048_12491213 | 0.16 |
ENSG00000223321 |
. |
84117 |
0.11 |
| chr11_12699031_12699547 | 0.16 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
2239 |
0.43 |
| chr6_86114788_86115158 | 0.16 |
NT5E |
5'-nucleotidase, ecto (CD73) |
44836 |
0.19 |
| chr3_127454933_127455119 | 0.16 |
MGLL |
monoglyceride lipase |
174 |
0.96 |
| chr11_89113858_89114276 | 0.16 |
NOX4 |
NADPH oxidase 4 |
109833 |
0.07 |
| chr18_43061735_43061886 | 0.16 |
RP11-309E23.2 |
|
1373 |
0.35 |
| chr4_81195528_81195918 | 0.16 |
FGF5 |
fibroblast growth factor 5 |
7930 |
0.26 |
| chr2_163098757_163099006 | 0.16 |
FAP |
fibroblast activation protein, alpha |
677 |
0.75 |
| chr5_54456050_54456451 | 0.16 |
GPX8 |
glutathione peroxidase 8 (putative) |
252 |
0.86 |
| chr4_41215270_41215769 | 0.16 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
956 |
0.59 |
| chr4_128553582_128553997 | 0.16 |
INTU |
inturned planar cell polarity protein |
298 |
0.94 |
| chr21_17654570_17655007 | 0.16 |
ENSG00000201025 |
. |
2301 |
0.46 |
| chr10_128110723_128110922 | 0.16 |
ADAM12 |
ADAM metallopeptidase domain 12 |
33798 |
0.19 |
| chr5_149534487_149534742 | 0.15 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
576 |
0.7 |
| chr12_106530238_106530817 | 0.15 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
3284 |
0.3 |
| chr2_166326382_166326565 | 0.15 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
265 |
0.96 |
| chr13_45147711_45147927 | 0.15 |
TSC22D1 |
TSC22 domain family, member 1 |
2573 |
0.38 |
| chr1_172109829_172110023 | 0.15 |
ENSG00000207949 |
. |
1879 |
0.31 |
| chr8_93074323_93074520 | 0.15 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
770 |
0.78 |
| chr1_172112919_172113144 | 0.15 |
ENSG00000208024 |
. |
753 |
0.46 |
| chr9_18475768_18475931 | 0.15 |
ADAMTSL1 |
ADAMTS-like 1 |
1618 |
0.53 |
| chr11_121969171_121969347 | 0.15 |
ENSG00000207971 |
. |
1293 |
0.36 |
| chr6_19691587_19692480 | 0.15 |
ENSG00000200957 |
. |
49273 |
0.18 |
| chr3_150088154_150088521 | 0.15 |
TSC22D2 |
TSC22 domain family, member 2 |
37785 |
0.2 |
| chr1_86047684_86048329 | 0.15 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
1562 |
0.37 |
| chr7_41740779_41741576 | 0.15 |
INHBA |
inhibin, beta A |
970 |
0.56 |
| chr6_169653019_169653284 | 0.15 |
THBS2 |
thrombospondin 2 |
905 |
0.7 |
| chr4_152146952_152147499 | 0.15 |
SH3D19 |
SH3 domain containing 19 |
7 |
0.98 |
| chr6_148664293_148664728 | 0.15 |
SASH1 |
SAM and SH3 domain containing 1 |
781 |
0.74 |
| chr7_116167346_116167524 | 0.15 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
1088 |
0.46 |
| chr8_17353947_17354148 | 0.15 |
SLC7A2 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
550 |
0.83 |
| chr5_54456456_54456818 | 0.15 |
GPX8 |
glutathione peroxidase 8 (putative) |
639 |
0.58 |
| chr12_13350846_13351208 | 0.15 |
EMP1 |
epithelial membrane protein 1 |
1307 |
0.55 |
| chr10_95241236_95241428 | 0.15 |
MYOF |
myoferlin |
619 |
0.72 |
| chr12_54017495_54017675 | 0.15 |
ATF7 |
activating transcription factor 7 |
2170 |
0.23 |
| chr3_168759646_168760009 | 0.15 |
MECOM |
MDS1 and EVI1 complex locus |
85995 |
0.11 |
| chr1_95005421_95005705 | 0.15 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
1630 |
0.52 |
| chr17_1665908_1666244 | 0.14 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
47 |
0.96 |
| chr2_98765349_98765752 | 0.14 |
VWA3B |
von Willebrand factor A domain containing 3B |
61851 |
0.13 |
| chr5_121305560_121305763 | 0.14 |
SRFBP1 |
serum response factor binding protein 1 |
8005 |
0.28 |
| chr7_94027358_94027511 | 0.14 |
COL1A2 |
collagen, type I, alpha 2 |
3561 |
0.34 |
| chr12_960802_961189 | 0.14 |
WNK1 |
WNK lysine deficient protein kinase 1 |
136 |
0.97 |
| chr1_61586045_61586196 | 0.14 |
RP4-802A10.1 |
|
4285 |
0.26 |
| chr13_24045503_24045654 | 0.14 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
37737 |
0.21 |
| chr3_55520495_55520928 | 0.14 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
620 |
0.62 |
| chr8_55294341_55294557 | 0.14 |
ENSG00000244107 |
. |
38579 |
0.17 |
| chr7_28111372_28111552 | 0.14 |
JAZF1 |
JAZF zinc finger 1 |
159 |
0.97 |
| chr11_121970307_121971111 | 0.14 |
ENSG00000207971 |
. |
157 |
0.79 |
| chr15_49715580_49715731 | 0.14 |
FGF7 |
fibroblast growth factor 7 |
198 |
0.96 |
| chr5_158176993_158177383 | 0.14 |
CTD-2363C16.1 |
|
232826 |
0.02 |
| chr4_144208077_144208509 | 0.14 |
ENSG00000200049 |
. |
22326 |
0.19 |
| chr2_190043558_190043796 | 0.14 |
COL5A2 |
collagen, type V, alpha 2 |
928 |
0.65 |
| chr18_56245541_56245920 | 0.14 |
RP11-126O1.2 |
|
21428 |
0.14 |
| chr7_100770539_100770851 | 0.14 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
316 |
0.81 |
| chr4_119811163_119811314 | 0.14 |
SYNPO2 |
synaptopodin 2 |
1046 |
0.64 |
| chr3_100785098_100785499 | 0.14 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
72939 |
0.11 |
| chr10_15667073_15667371 | 0.14 |
ITGA8 |
integrin, alpha 8 |
94902 |
0.09 |
| chr5_72733592_72733937 | 0.14 |
FOXD1 |
forkhead box D1 |
10588 |
0.21 |
| chr5_57753977_57754128 | 0.14 |
PLK2 |
polo-like kinase 2 |
2035 |
0.38 |
| chr2_190044119_190044339 | 0.14 |
COL5A2 |
collagen, type V, alpha 2 |
376 |
0.9 |
| chr2_238318512_238318719 | 0.14 |
COL6A3 |
collagen, type VI, alpha 3 |
4176 |
0.24 |
| chr4_23890359_23890765 | 0.14 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
1096 |
0.66 |
| chr2_197831667_197831951 | 0.14 |
ANKRD44 |
ankyrin repeat domain 44 |
33417 |
0.18 |
| chr15_63673315_63673584 | 0.14 |
CA12 |
carbonic anhydrase XII |
585 |
0.82 |
| chrX_3467246_3467792 | 0.14 |
ENSG00000212214 |
. |
17227 |
0.19 |
| chr5_38555642_38555941 | 0.14 |
LIFR |
leukemia inhibitory factor receptor alpha |
947 |
0.37 |
| chr4_162267666_162267817 | 0.14 |
RP11-234O6.2 |
|
32364 |
0.26 |
| chr3_15838336_15838568 | 0.14 |
ANKRD28 |
ankyrin repeat domain 28 |
424 |
0.86 |
| chr11_69458516_69459047 | 0.14 |
CCND1 |
cyclin D1 |
2807 |
0.29 |
| chr8_92612283_92612746 | 0.14 |
ENSG00000200151 |
. |
5321 |
0.33 |
| chr7_18549761_18549974 | 0.14 |
HDAC9 |
histone deacetylase 9 |
931 |
0.71 |
| chr18_416158_416713 | 0.13 |
RP11-720L2.2 |
|
7981 |
0.25 |
| chr3_55520023_55520469 | 0.13 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1085 |
0.48 |
| chr12_91575190_91575391 | 0.13 |
DCN |
decorin |
537 |
0.85 |
| chr1_214723918_214724069 | 0.13 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
573 |
0.84 |
| chr4_151500686_151500914 | 0.13 |
RP11-1336O20.2 |
|
559 |
0.76 |
| chr7_41742004_41742175 | 0.13 |
INHBA |
inhibin, beta A |
617 |
0.72 |
| chr2_164591967_164592245 | 0.13 |
FIGN |
fidgetin |
411 |
0.92 |
| chr10_33622034_33622426 | 0.13 |
NRP1 |
neuropilin 1 |
1080 |
0.63 |
| chr10_35102663_35102949 | 0.13 |
PARD3 |
par-3 family cell polarity regulator |
1443 |
0.4 |
| chr15_37394480_37395011 | 0.13 |
MEIS2 |
Meis homeobox 2 |
1241 |
0.49 |
| chr3_66139685_66140118 | 0.13 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
20616 |
0.25 |
| chr10_95239483_95239707 | 0.13 |
MYOF |
myoferlin |
2356 |
0.29 |
| chr15_49718765_49718970 | 0.13 |
FGF7 |
fibroblast growth factor 7 |
3410 |
0.29 |
| chr22_45865037_45865472 | 0.13 |
RP1-102D24.5 |
|
20630 |
0.17 |
| chr16_21199797_21200120 | 0.13 |
AF001550.7 |
|
6210 |
0.15 |
| chr19_18497517_18497681 | 0.13 |
ENSG00000264175 |
. |
227 |
0.74 |
| chr4_23890131_23890306 | 0.13 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
1440 |
0.57 |
| chr19_47735595_47736220 | 0.13 |
BBC3 |
BCL2 binding component 3 |
116 |
0.95 |
| chr21_28217168_28217584 | 0.13 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
352 |
0.91 |
| chr5_40552090_40552408 | 0.13 |
ENSG00000199552 |
. |
102816 |
0.07 |
| chr3_190580567_190581193 | 0.13 |
GMNC |
geminin coiled-coil domain containing |
476 |
0.9 |
| chr11_101983358_101983513 | 0.13 |
YAP1 |
Yes-associated protein 1 |
190 |
0.94 |
| chr13_38171648_38172094 | 0.13 |
POSTN |
periostin, osteoblast specific factor |
992 |
0.71 |
| chr12_69418432_69418757 | 0.13 |
CPM |
carboxypeptidase M |
61545 |
0.11 |
| chr12_13351879_13352395 | 0.13 |
EMP1 |
epithelial membrane protein 1 |
2417 |
0.37 |
| chr3_99356455_99356606 | 0.13 |
COL8A1 |
collagen, type VIII, alpha 1 |
789 |
0.74 |
| chr12_104982084_104982267 | 0.13 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
451 |
0.85 |
| chr1_216773992_216774229 | 0.13 |
ESRRG |
estrogen-related receptor gamma |
122564 |
0.06 |
| chr3_11196334_11196700 | 0.13 |
HRH1 |
histamine receptor H1 |
303 |
0.94 |
| chr11_57529774_57530544 | 0.13 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
869 |
0.5 |
| chr6_56556821_56557065 | 0.13 |
DST |
dystonin |
49149 |
0.18 |
| chr12_115119174_115119325 | 0.13 |
TBX3 |
T-box 3 |
2146 |
0.36 |
| chr4_111557687_111558154 | 0.13 |
PITX2 |
paired-like homeodomain 2 |
256 |
0.95 |
| chr2_163099691_163099877 | 0.13 |
FAP |
fibroblast activation protein, alpha |
182 |
0.96 |
| chr2_67601751_67601914 | 0.13 |
ETAA1 |
Ewing tumor-associated antigen 1 |
22619 |
0.29 |
| chr18_52626293_52626952 | 0.13 |
CCDC68 |
coiled-coil domain containing 68 |
117 |
0.97 |
| chr10_111765795_111766267 | 0.13 |
ADD3 |
adducin 3 (gamma) |
469 |
0.79 |
| chr1_182931948_182932121 | 0.13 |
ENSG00000264768 |
. |
3324 |
0.2 |
| chr14_52121858_52122101 | 0.13 |
FRMD6 |
FERM domain containing 6 |
3281 |
0.25 |
| chr8_27694457_27694746 | 0.13 |
PBK |
PDZ binding kinase |
998 |
0.51 |
| chr2_235862821_235863118 | 0.13 |
SH3BP4 |
SH3-domain binding protein 4 |
2249 |
0.48 |
| chr2_74780571_74780754 | 0.13 |
LOXL3 |
lysyl oxidase-like 3 |
400 |
0.5 |
| chr11_95835777_95835928 | 0.13 |
MTMR2 |
myotubularin related protein 2 |
178393 |
0.03 |
| chr3_114790362_114790748 | 0.13 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
333 |
0.94 |
| chr11_79148844_79149050 | 0.13 |
TENM4 |
teneurin transmembrane protein 4 |
2748 |
0.3 |
| chr22_38864455_38864654 | 0.13 |
KDELR3 |
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
471 |
0.74 |
| chr5_124536368_124536614 | 0.13 |
ENSG00000222107 |
. |
150333 |
0.05 |
| chr15_63337269_63337696 | 0.13 |
TPM1 |
tropomyosin 1 (alpha) |
1507 |
0.37 |
| chr6_56557534_56557976 | 0.13 |
DST |
dystonin |
49961 |
0.18 |
| chr7_23286622_23287130 | 0.13 |
GPNMB |
glycoprotein (transmembrane) nmb |
482 |
0.83 |
| chr1_170634717_170634868 | 0.13 |
PRRX1 |
paired related homeobox 1 |
1714 |
0.5 |
| chr3_124604600_124604823 | 0.12 |
ITGB5 |
integrin, beta 5 |
1433 |
0.47 |
| chr5_54457024_54457471 | 0.12 |
GPX8 |
glutathione peroxidase 8 (putative) |
1249 |
0.31 |
| chr7_130595370_130595521 | 0.12 |
ENSG00000226380 |
. |
33147 |
0.2 |
| chr9_13787422_13787665 | 0.12 |
NFIB |
nuclear factor I/B |
393248 |
0.01 |
| chr18_73688861_73689088 | 0.12 |
RP11-94B19.4 |
Uncharacterized protein |
282147 |
0.01 |
| chr11_133995025_133995278 | 0.12 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
43216 |
0.13 |
| chr14_27065632_27065895 | 0.12 |
NOVA1 |
neuro-oncological ventral antigen 1 |
482 |
0.74 |
| chr7_27218725_27218915 | 0.12 |
RP1-170O19.20 |
Uncharacterized protein |
812 |
0.27 |
| chr3_29325145_29325351 | 0.12 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
2141 |
0.37 |
| chr21_17442999_17443232 | 0.12 |
ENSG00000252273 |
. |
35286 |
0.24 |
| chr10_4285552_4285907 | 0.12 |
ENSG00000207124 |
. |
271415 |
0.02 |
| chr7_55088709_55088860 | 0.12 |
EGFR |
epidermal growth factor receptor |
1973 |
0.48 |
| chr9_22084576_22084735 | 0.12 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
29022 |
0.19 |
| chr13_35963159_35963423 | 0.12 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
87541 |
0.09 |
| chr15_56206351_56206878 | 0.12 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
1238 |
0.55 |
| chr14_73492682_73492833 | 0.12 |
ZFYVE1 |
zinc finger, FYVE domain containing 1 |
1057 |
0.51 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.2 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.3 | GO:0097435 | extracellular fibril organization(GO:0043206) fibril organization(GO:0097435) |
| 0.1 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.1 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.4 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.3 | GO:0055093 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.2 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.2 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0060363 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.2 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.1 | GO:0090196 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0015802 | basic amino acid transport(GO:0015802) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0010667 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:2000108 | positive regulation of leukocyte apoptotic process(GO:2000108) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0009629 | response to gravity(GO:0009629) |
| 0.0 | 0.0 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.3 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.0 | GO:0051280 | negative regulation of ion transmembrane transporter activity(GO:0032413) negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.0 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.2 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0060214 | endocardium development(GO:0003157) endocardium morphogenesis(GO:0003160) endocardium formation(GO:0060214) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0002913 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.3 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.1 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.0 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.0 | GO:1903513 | retrograde protein transport, ER to cytosol(GO:0030970) endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 0.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.0 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.1 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.2 | 0.5 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.7 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.3 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.0 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.3 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.0 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME PI 3K CASCADE | Genes involved in PI-3K cascade |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |