Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
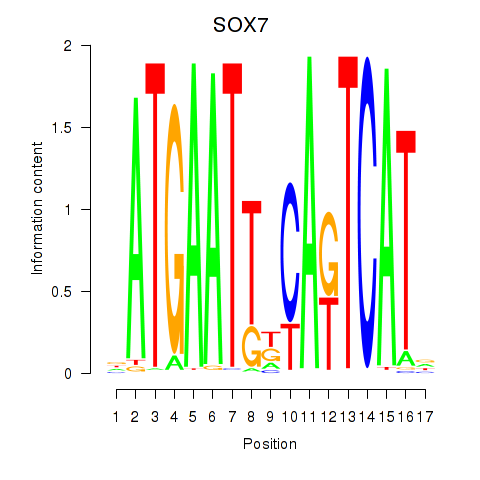
Results for SOX7
Z-value: 0.70
Transcription factors associated with SOX7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX7
|
ENSG00000171056.6 | SRY-box transcription factor 7 |
|
SOX7
|
ENSG00000258724.1 | SRY-box transcription factor 7 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_10588268_10588485 | SOX7 | 354 | 0.784535 | 0.65 | 5.6e-02 | Click! |
| chr8_10591974_10592215 | SOX7 | 4072 | 0.173015 | -0.48 | 1.9e-01 | Click! |
| chr8_10593745_10593896 | SOX7 | 5798 | 0.158950 | 0.47 | 2.0e-01 | Click! |
| chr8_10590828_10591128 | SOX7 | 2956 | 0.196224 | 0.42 | 2.6e-01 | Click! |
| chr8_10591581_10591861 | SOX7 | 3699 | 0.178656 | 0.31 | 4.1e-01 | Click! |
Activity of the SOX7 motif across conditions
Conditions sorted by the z-value of the SOX7 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
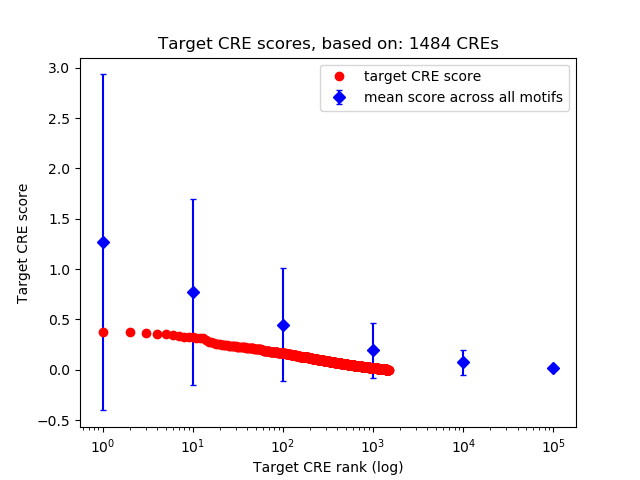
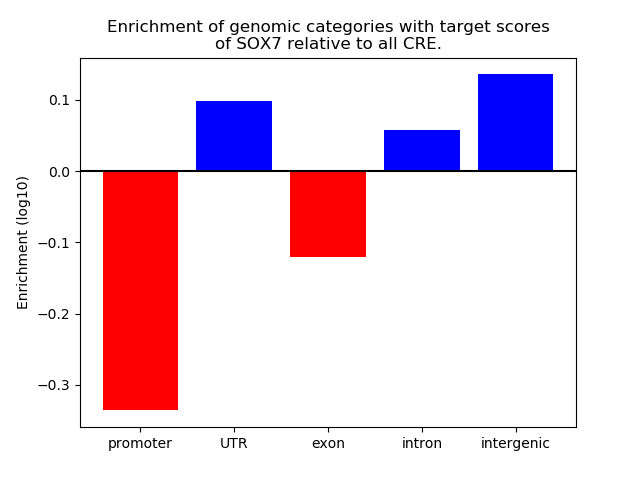
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_28997424_28997626 | 0.38 |
FLT1 |
fms-related tyrosine kinase 1 |
71707 |
0.12 |
| chr14_37570606_37570757 | 0.37 |
RP11-537P22.2 |
|
2913 |
0.31 |
| chr2_102858447_102858598 | 0.36 |
IL1RL2 |
interleukin 1 receptor-like 2 |
54748 |
0.11 |
| chr1_14032622_14032789 | 0.36 |
PRDM2 |
PR domain containing 2, with ZNF domain |
1355 |
0.46 |
| chr5_179495571_179495863 | 0.36 |
RNF130 |
ring finger protein 130 |
3004 |
0.28 |
| chr11_123349536_123349687 | 0.35 |
GRAMD1B |
GRAM domain containing 1B |
46733 |
0.14 |
| chr11_14375529_14375805 | 0.34 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
379 |
0.91 |
| chr7_9336486_9336637 | 0.33 |
ENSG00000264766 |
. |
297792 |
0.01 |
| chr13_41579962_41580113 | 0.33 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
13413 |
0.19 |
| chr5_85380672_85380823 | 0.32 |
NBPF22P |
neuroblastoma breakpoint family, member 22, pseudogene |
197837 |
0.03 |
| chr2_169962924_169963075 | 0.32 |
AC007556.3 |
|
5746 |
0.24 |
| chr4_10182968_10183393 | 0.32 |
WDR1 |
WD repeat domain 1 |
64607 |
0.11 |
| chr6_14721538_14721689 | 0.32 |
ENSG00000206960 |
. |
74847 |
0.13 |
| chr13_74185476_74185627 | 0.29 |
KLF12 |
Kruppel-like factor 12 |
383635 |
0.01 |
| chr1_76788853_76789004 | 0.28 |
ST6GALNAC3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
248524 |
0.02 |
| chr1_72809770_72809921 | 0.27 |
NEGR1 |
neuronal growth regulator 1 |
61428 |
0.16 |
| chr5_57555667_57556212 | 0.27 |
ENSG00000238899 |
. |
189924 |
0.03 |
| chr4_184509430_184509787 | 0.26 |
ENSG00000239116 |
. |
6154 |
0.2 |
| chr1_169454193_169456202 | 0.26 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
28 |
0.98 |
| chr2_200604849_200605000 | 0.25 |
FTCDNL1 |
formiminotransferase cyclodeaminase N-terminal like |
110672 |
0.07 |
| chr1_155730342_155730493 | 0.25 |
DAP3 |
death associated protein 3 |
23614 |
0.14 |
| chr6_42781709_42781860 | 0.25 |
GLTSCR1L |
GLTSCR1-like |
7010 |
0.18 |
| chr15_83861368_83861519 | 0.25 |
RP11-382A20.4 |
|
7886 |
0.18 |
| chr2_121015272_121015465 | 0.24 |
RALB |
v-ral simian leukemia viral oncogene homolog B |
4341 |
0.23 |
| chr18_56345553_56345704 | 0.24 |
RP11-126O1.4 |
|
782 |
0.57 |
| chr3_185478819_185478970 | 0.24 |
ENSG00000265470 |
. |
6798 |
0.24 |
| chr4_99829065_99829216 | 0.24 |
RP11-571L19.7 |
|
20908 |
0.15 |
| chr5_43307969_43308120 | 0.24 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
5282 |
0.22 |
| chr12_66198930_66199081 | 0.23 |
HMGA2 |
high mobility group AT-hook 2 |
18906 |
0.21 |
| chr5_52293053_52293204 | 0.23 |
CTD-2175A23.1 |
|
7020 |
0.22 |
| chr3_74315841_74315992 | 0.23 |
ENSG00000212585 |
. |
100713 |
0.09 |
| chrX_27636247_27636398 | 0.23 |
DCAF8L2 |
DDB1 and CUL4 associated factor 8-like 2 |
27805 |
0.27 |
| chr2_145195065_145195216 | 0.23 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
7003 |
0.3 |
| chr22_31369216_31369367 | 0.23 |
TUG1 |
taurine up-regulated 1 (non-protein coding) |
107 |
0.96 |
| chr5_107195765_107195916 | 0.23 |
ENSG00000251732 |
. |
49510 |
0.19 |
| chr4_184922100_184922251 | 0.23 |
STOX2 |
storkhead box 2 |
303 |
0.92 |
| chr2_64102829_64103045 | 0.23 |
ENSG00000251775 |
. |
7588 |
0.24 |
| chr20_14316551_14316865 | 0.22 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
1546 |
0.47 |
| chr2_124669235_124669386 | 0.22 |
ENSG00000223118 |
. |
42163 |
0.2 |
| chr8_10268503_10268654 | 0.22 |
RP11-981G7.1 |
|
22604 |
0.2 |
| chr4_95455492_95455643 | 0.22 |
PDLIM5 |
PDZ and LIM domain 5 |
10691 |
0.32 |
| chr12_129315092_129315280 | 0.22 |
SLC15A4 |
solute carrier family 15 (oligopeptide transporter), member 4 |
6658 |
0.26 |
| chr5_71543858_71544009 | 0.22 |
ENSG00000244748 |
. |
66582 |
0.09 |
| chr13_107166471_107166622 | 0.22 |
EFNB2 |
ephrin-B2 |
20916 |
0.25 |
| chr3_60921693_60921844 | 0.21 |
ENSG00000212211 |
. |
79491 |
0.12 |
| chr6_27109416_27109567 | 0.21 |
HIST1H4I |
histone cluster 1, H4i |
2415 |
0.16 |
| chr1_234659025_234659394 | 0.21 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
44360 |
0.14 |
| chr2_46294741_46294968 | 0.21 |
AC017006.2 |
|
11113 |
0.26 |
| chr10_49573800_49573951 | 0.21 |
MAPK8 |
mitogen-activated protein kinase 8 |
35811 |
0.19 |
| chr6_142315032_142315183 | 0.21 |
RP11-137J7.2 |
|
94263 |
0.08 |
| chr6_109397933_109398084 | 0.21 |
ENSG00000199944 |
. |
17191 |
0.17 |
| chr7_116295969_116296120 | 0.21 |
MET |
met proto-oncogene |
16400 |
0.22 |
| chrX_46509501_46509746 | 0.21 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
76404 |
0.1 |
| chr5_177609718_177609869 | 0.21 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
21715 |
0.15 |
| chr8_119669481_119669632 | 0.20 |
SAMD12 |
sterile alpha motif domain containing 12 |
35322 |
0.24 |
| chr1_108115540_108115852 | 0.20 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
115430 |
0.06 |
| chr2_227670394_227670545 | 0.20 |
RP11-395N3.1 |
|
1717 |
0.38 |
| chr21_30580723_30581000 | 0.20 |
BACH1 |
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
14393 |
0.13 |
| chr12_67476278_67476429 | 0.20 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
186708 |
0.03 |
| chr12_66349719_66349870 | 0.19 |
RP11-366L20.3 |
|
7535 |
0.21 |
| chr1_200417818_200417969 | 0.19 |
ZNF281 |
zinc finger protein 281 |
38709 |
0.21 |
| chr6_106382736_106382887 | 0.19 |
ENSG00000200198 |
. |
30563 |
0.24 |
| chr11_93868067_93868416 | 0.19 |
PANX1 |
pannexin 1 |
6146 |
0.28 |
| chr21_30007689_30007840 | 0.19 |
ENSG00000251894 |
. |
107766 |
0.07 |
| chr16_2652914_2653065 | 0.19 |
AC141586.5 |
|
486 |
0.59 |
| chr9_110011277_110011428 | 0.19 |
RAD23B |
RAD23 homolog B (S. cerevisiae) |
34066 |
0.22 |
| chr7_46794308_46794459 | 0.18 |
AC011294.3 |
Uncharacterized protein |
57663 |
0.17 |
| chr1_86034185_86034336 | 0.18 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
9673 |
0.19 |
| chr8_26175473_26175624 | 0.18 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
24549 |
0.21 |
| chr7_32102480_32102631 | 0.18 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
7906 |
0.33 |
| chr7_24576141_24576292 | 0.18 |
ENSG00000206877 |
. |
735 |
0.75 |
| chr12_60541434_60541585 | 0.18 |
ENSG00000264437 |
. |
120871 |
0.06 |
| chr3_186475417_186475568 | 0.18 |
RP11-573D15.8 |
|
35 |
0.94 |
| chr15_40399291_40399456 | 0.18 |
BMF |
Bcl2 modifying factor |
527 |
0.74 |
| chrX_45482024_45482567 | 0.18 |
ENSG00000207870 |
. |
123399 |
0.06 |
| chr5_124341116_124341267 | 0.18 |
ZNF608 |
zinc finger protein 608 |
256691 |
0.02 |
| chr17_69917748_69917899 | 0.18 |
AC007461.1 |
Uncharacterized protein |
118341 |
0.07 |
| chr2_113593759_113594336 | 0.18 |
IL1B |
interleukin 1, beta |
36 |
0.97 |
| chr8_57964344_57964495 | 0.18 |
IMPAD1 |
inositol monophosphatase domain containing 1 |
58016 |
0.14 |
| chr1_234738398_234739129 | 0.18 |
RP4-781K5.2 |
|
3991 |
0.21 |
| chr21_37445377_37445528 | 0.18 |
AP000688.14 |
|
2448 |
0.16 |
| chr12_104766180_104766331 | 0.18 |
TXNRD1 |
thioredoxin reductase 1 |
62626 |
0.11 |
| chr5_131717278_131717429 | 0.18 |
SLC22A5 |
solute carrier family 22 (organic cation/carnitine transporter), member 5 |
11385 |
0.12 |
| chr5_57758229_57758380 | 0.18 |
PLK2 |
polo-like kinase 2 |
2217 |
0.35 |
| chr16_87469821_87470025 | 0.18 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
3183 |
0.21 |
| chr5_39076739_39076970 | 0.18 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
2344 |
0.37 |
| chr3_18763882_18764033 | 0.17 |
ENSG00000228956 |
. |
23279 |
0.28 |
| chr2_143632357_143632508 | 0.17 |
KYNU |
kynureninase |
2635 |
0.42 |
| chr2_134575536_134575687 | 0.17 |
ENSG00000200708 |
. |
221949 |
0.02 |
| chr2_235170388_235170539 | 0.17 |
SPP2 |
secreted phosphoprotein 2, 24kDa |
211032 |
0.02 |
| chr6_113614101_113614252 | 0.17 |
ENSG00000222677 |
. |
222013 |
0.02 |
| chr3_188302556_188302930 | 0.17 |
LPP-AS1 |
LPP antisense RNA 1 |
16289 |
0.2 |
| chr5_84011446_84011597 | 0.17 |
CTD-2269F5.1 |
|
330224 |
0.01 |
| chr20_50417404_50418104 | 0.17 |
SALL4 |
spalt-like transcription factor 4 |
1193 |
0.55 |
| chr5_106724870_106725112 | 0.17 |
EFNA5 |
ephrin-A5 |
281337 |
0.01 |
| chr10_5652217_5652368 | 0.17 |
ENSG00000240577 |
. |
21134 |
0.16 |
| chr16_86619409_86619937 | 0.17 |
FOXL1 |
forkhead box L1 |
7558 |
0.18 |
| chr12_51008245_51008396 | 0.17 |
ENSG00000207136 |
. |
18736 |
0.2 |
| chr9_127999236_127999759 | 0.16 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
4112 |
0.17 |
| chr1_226229292_226229443 | 0.16 |
H3F3A |
H3 histone, family 3A |
20185 |
0.15 |
| chr8_92050598_92050749 | 0.16 |
TMEM55A |
transmembrane protein 55A |
2390 |
0.2 |
| chr6_86303657_86304481 | 0.16 |
SNX14 |
sorting nexin 14 |
195 |
0.95 |
| chr6_142632086_142632237 | 0.16 |
GPR126 |
G protein-coupled receptor 126 |
8398 |
0.27 |
| chr2_81431264_81431415 | 0.16 |
ENSG00000200890 |
. |
292112 |
0.01 |
| chr10_132429038_132429331 | 0.16 |
ENSG00000222997 |
. |
125814 |
0.06 |
| chr10_83364907_83365058 | 0.16 |
NRG3 |
neuregulin 3 |
270088 |
0.02 |
| chr1_78469337_78469488 | 0.16 |
RP11-386I14.4 |
|
826 |
0.45 |
| chr13_47181821_47181972 | 0.16 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
54501 |
0.16 |
| chr20_10502111_10502382 | 0.16 |
SLX4IP |
SLX4 interacting protein |
86295 |
0.09 |
| chr5_143062873_143063024 | 0.16 |
ENSG00000266478 |
. |
3523 |
0.34 |
| chr2_144704936_144705087 | 0.16 |
AC016910.1 |
|
10371 |
0.27 |
| chr1_175289119_175289270 | 0.16 |
RP3-518E13.2 |
|
12840 |
0.27 |
| chrX_108952305_108952710 | 0.16 |
ACSL4 |
acyl-CoA synthetase long-chain family member 4 |
17131 |
0.22 |
| chr21_20465237_20465388 | 0.16 |
ENSG00000212609 |
. |
252153 |
0.02 |
| chr21_39869780_39870199 | 0.16 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
356 |
0.93 |
| chr5_98269739_98269890 | 0.15 |
ENSG00000200351 |
. |
2637 |
0.3 |
| chr12_54676753_54676980 | 0.15 |
HNRNPA1 |
heterogeneous nuclear ribonucleoprotein A1 |
884 |
0.34 |
| chr14_81859528_81859679 | 0.15 |
STON2 |
stonin 2 |
5324 |
0.33 |
| chr6_108910719_108911119 | 0.15 |
FOXO3 |
forkhead box O3 |
28850 |
0.24 |
| chr9_97568099_97568250 | 0.15 |
ENSG00000252153 |
. |
4070 |
0.21 |
| chr1_36104081_36104260 | 0.15 |
PSMB2 |
proteasome (prosome, macropain) subunit, beta type, 2 |
3275 |
0.22 |
| chr18_48571368_48571694 | 0.15 |
SMAD4 |
SMAD family member 4 |
202 |
0.96 |
| chr12_51328679_51328941 | 0.15 |
ENSG00000199740 |
. |
4999 |
0.16 |
| chr2_157748866_157749017 | 0.15 |
ENSG00000263848 |
. |
122828 |
0.06 |
| chr6_147438064_147438215 | 0.15 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
2460 |
0.43 |
| chr3_58481014_58481165 | 0.15 |
KCTD6 |
potassium channel tetramerization domain containing 6 |
1447 |
0.4 |
| chr1_110138366_110138517 | 0.15 |
RP5-1160K1.3 |
|
406 |
0.68 |
| chr10_112624321_112624902 | 0.15 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
6954 |
0.15 |
| chr10_3902959_3903110 | 0.15 |
KLF6 |
Kruppel-like factor 6 |
75561 |
0.11 |
| chr5_95360695_95360846 | 0.15 |
ENSG00000207578 |
. |
54072 |
0.12 |
| chr2_47194995_47195171 | 0.14 |
RP11-15I20.1 |
|
11056 |
0.16 |
| chr2_12199767_12199918 | 0.14 |
ENSG00000238503 |
. |
29344 |
0.24 |
| chr2_113065761_113065912 | 0.14 |
ZC3H6 |
zinc finger CCCH-type containing 6 |
32658 |
0.16 |
| chr10_60400935_60401183 | 0.14 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
128159 |
0.06 |
| chr5_39415323_39415474 | 0.14 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
9572 |
0.27 |
| chr14_91882732_91883366 | 0.14 |
CCDC88C |
coiled-coil domain containing 88C |
641 |
0.78 |
| chr3_85210330_85210481 | 0.14 |
CADM2 |
cell adhesion molecule 2 |
201708 |
0.03 |
| chr5_145318270_145318548 | 0.14 |
SH3RF2 |
SH3 domain containing ring finger 2 |
969 |
0.66 |
| chr5_131798996_131799147 | 0.14 |
ENSG00000202533 |
. |
4768 |
0.15 |
| chr6_10401153_10401304 | 0.14 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
3599 |
0.23 |
| chr15_94406450_94406737 | 0.14 |
ENSG00000222409 |
. |
33663 |
0.25 |
| chr11_100586653_100586804 | 0.14 |
CTD-2383M3.1 |
|
28042 |
0.18 |
| chr6_36187187_36187338 | 0.14 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
15071 |
0.15 |
| chr1_81929626_81929777 | 0.14 |
RP5-837I24.6 |
|
41083 |
0.2 |
| chr4_165110997_165111148 | 0.14 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
194069 |
0.03 |
| chr9_80595790_80595941 | 0.14 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
49655 |
0.19 |
| chr13_73664940_73665091 | 0.14 |
ENSG00000200267 |
. |
22140 |
0.18 |
| chr3_194026692_194026843 | 0.14 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
45280 |
0.14 |
| chr1_246772402_246772553 | 0.14 |
RP11-439E19.1 |
|
3302 |
0.26 |
| chr9_76788506_76788657 | 0.14 |
RORB |
RAR-related orphan receptor B |
323700 |
0.01 |
| chr5_30104022_30104173 | 0.14 |
ENSG00000223136 |
. |
155162 |
0.05 |
| chr7_25902676_25902879 | 0.14 |
ENSG00000199085 |
. |
86829 |
0.1 |
| chr21_34587468_34587880 | 0.13 |
IFNAR2 |
interferon (alpha, beta and omega) receptor 2 |
14532 |
0.14 |
| chr12_102454924_102455862 | 0.13 |
CCDC53 |
coiled-coil domain containing 53 |
464 |
0.76 |
| chr4_47914207_47914358 | 0.13 |
NFXL1 |
nuclear transcription factor, X-box binding-like 1 |
2274 |
0.31 |
| chr4_110361887_110362038 | 0.13 |
SEC24B-AS1 |
SEC24B antisense RNA 1 |
6989 |
0.17 |
| chr5_147778011_147778162 | 0.13 |
FBXO38 |
F-box protein 38 |
3811 |
0.2 |
| chr11_18640098_18640349 | 0.13 |
SPTY2D1 |
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
15817 |
0.14 |
| chr1_182937815_182937966 | 0.13 |
ENSG00000264768 |
. |
9180 |
0.15 |
| chr14_94346395_94346670 | 0.13 |
FAM181A |
family with sequence similarity 181, member A |
38708 |
0.14 |
| chr2_192217267_192217418 | 0.13 |
ENSG00000252130 |
. |
18322 |
0.24 |
| chr3_43601794_43601945 | 0.13 |
ANO10 |
anoctamin 10 |
4966 |
0.3 |
| chr15_76091845_76091996 | 0.13 |
ENSG00000241807 |
. |
14467 |
0.11 |
| chr1_206138782_206139735 | 0.13 |
FAM72A |
family with sequence similarity 72, member A |
347 |
0.82 |
| chr17_5513967_5514118 | 0.13 |
NLRP1 |
NLR family, pyrin domain containing 1 |
8702 |
0.24 |
| chr7_125201444_125201595 | 0.13 |
ENSG00000221418 |
. |
478764 |
0.01 |
| chr6_20497460_20497742 | 0.13 |
ENSG00000201683 |
. |
2805 |
0.26 |
| chr15_67533700_67533851 | 0.13 |
AAGAB |
alpha- and gamma-adaptin binding protein |
1434 |
0.45 |
| chr2_133998912_133999202 | 0.13 |
AC010890.1 |
|
23483 |
0.24 |
| chr3_9436532_9436683 | 0.13 |
SETD5-AS1 |
SETD5 antisense RNA 1 |
1682 |
0.28 |
| chr4_20196394_20196545 | 0.13 |
SLIT2 |
slit homolog 2 (Drosophila) |
58414 |
0.17 |
| chr8_62621525_62621728 | 0.13 |
ASPH |
aspartate beta-hydroxylase |
5458 |
0.25 |
| chr20_19961524_19961675 | 0.13 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
36161 |
0.17 |
| chr1_143911814_143912529 | 0.13 |
FAM72D |
family with sequence similarity 72, member D |
972 |
0.56 |
| chr5_32441809_32441985 | 0.13 |
ZFR |
zinc finger RNA binding protein |
2970 |
0.29 |
| chr10_129848775_129848926 | 0.13 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
3016 |
0.36 |
| chrX_131994519_131994670 | 0.13 |
HS6ST2 |
heparan sulfate 6-O-sulfotransferase 2 |
96750 |
0.08 |
| chr22_27562410_27562561 | 0.12 |
ENSG00000200443 |
. |
130635 |
0.06 |
| chr1_24476638_24476996 | 0.12 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
7206 |
0.17 |
| chr7_130680709_130680860 | 0.12 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
11916 |
0.25 |
| chr11_15991196_15991378 | 0.12 |
CTD-3096P4.1 |
|
53449 |
0.18 |
| chr10_31271779_31271930 | 0.12 |
ZNF438 |
zinc finger protein 438 |
1501 |
0.54 |
| chr15_70702879_70703030 | 0.12 |
ENSG00000200216 |
. |
217379 |
0.02 |
| chr12_79803274_79803425 | 0.12 |
ENSG00000221788 |
. |
9688 |
0.22 |
| chr1_79761398_79761549 | 0.12 |
ENSG00000266388 |
. |
160562 |
0.04 |
| chr6_70219788_70219939 | 0.12 |
ENSG00000265362 |
. |
111402 |
0.07 |
| chr21_39678468_39679041 | 0.12 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
9906 |
0.29 |
| chr3_141205962_141207110 | 0.12 |
RASA2 |
RAS p21 protein activator 2 |
645 |
0.76 |
| chr6_111924581_111924732 | 0.12 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
2418 |
0.25 |
| chr10_38120780_38120931 | 0.12 |
ENSG00000266800 |
. |
38 |
0.98 |
| chr1_108584009_108584286 | 0.12 |
ENSG00000264753 |
. |
22754 |
0.22 |
| chr10_60273912_60274168 | 0.12 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
1140 |
0.65 |
| chr2_236618011_236618251 | 0.12 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
35277 |
0.2 |
| chr2_190444828_190446170 | 0.12 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
114 |
0.98 |
| chr11_34998257_34998466 | 0.12 |
PDHX |
pyruvate dehydrogenase complex, component X |
970 |
0.61 |
| chr7_82434749_82434900 | 0.12 |
PCLO |
piccolo presynaptic cytomatrix protein |
145240 |
0.05 |
| chr1_146633619_146633770 | 0.12 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
10352 |
0.16 |
| chr6_109644101_109644252 | 0.12 |
ENSG00000201023 |
. |
17479 |
0.16 |
| chr9_71459077_71459228 | 0.12 |
RP11-203L2.4 |
|
961 |
0.65 |
| chr1_14039870_14040021 | 0.12 |
PRDM2 |
PR domain containing 2, with ZNF domain |
8595 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0055094 | response to lipoprotein particle(GO:0055094) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |