Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
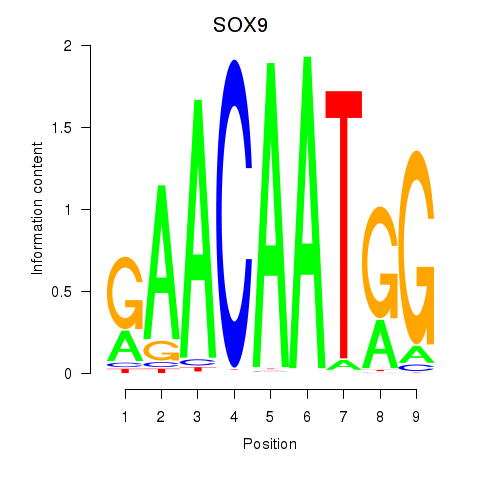
Results for SOX9
Z-value: 0.55
Transcription factors associated with SOX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SOX9
|
ENSG00000125398.5 | SRY-box transcription factor 9 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_70117705_70117946 | SOX9 | 664 | 0.821863 | -0.68 | 4.6e-02 | Click! |
| chr17_70118144_70118357 | SOX9 | 1089 | 0.670792 | -0.67 | 4.7e-02 | Click! |
| chr17_70358322_70358473 | SOX9 | 241236 | 0.019037 | -0.62 | 7.3e-02 | Click! |
| chr17_70113280_70113463 | SOX9 | 3790 | 0.358750 | -0.61 | 7.8e-02 | Click! |
| chr17_70269373_70269524 | SOX9 | 152287 | 0.043999 | -0.61 | 8.0e-02 | Click! |
Activity of the SOX9 motif across conditions
Conditions sorted by the z-value of the SOX9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
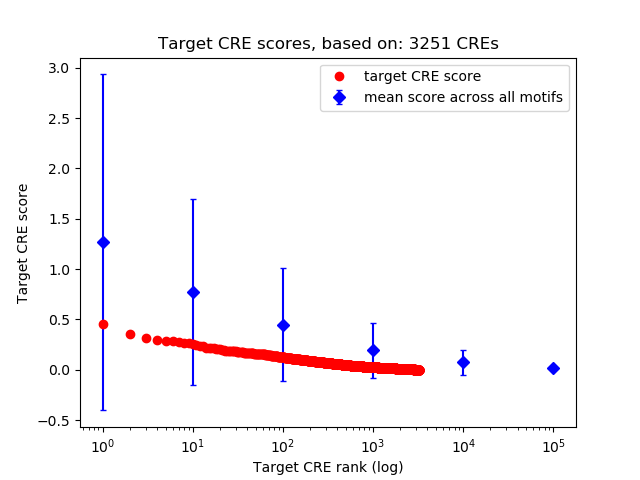
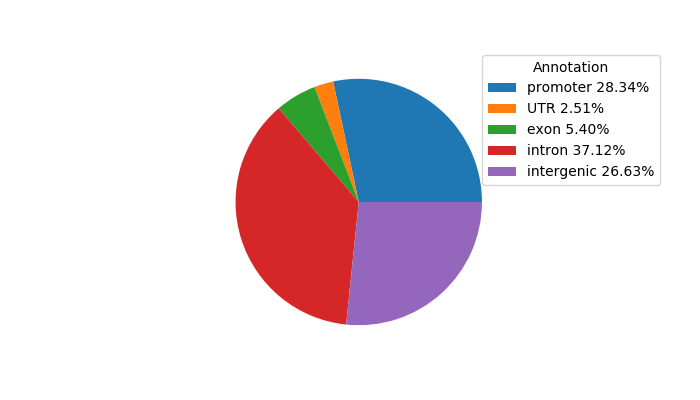
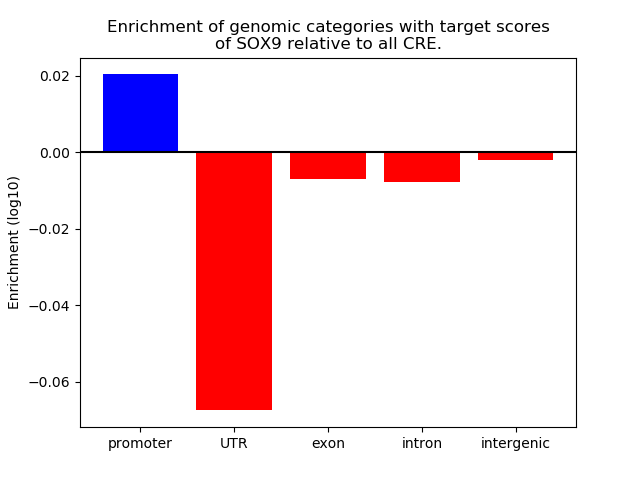
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr21_15917471_15917848 | 0.46 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
1003 |
0.63 |
| chr2_65245033_65245279 | 0.36 |
AC007386.4 |
|
19475 |
0.14 |
| chr15_86098904_86099149 | 0.31 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
349 |
0.88 |
| chr15_50411579_50411857 | 0.30 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
299 |
0.92 |
| chr1_212428535_212428756 | 0.29 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
30234 |
0.16 |
| chr12_10282570_10282865 | 0.29 |
CLEC7A |
C-type lectin domain family 7, member A |
35 |
0.96 |
| chr13_114823175_114823405 | 0.27 |
RASA3 |
RAS p21 protein activator 3 |
20148 |
0.23 |
| chr12_54891563_54891796 | 0.27 |
NCKAP1L |
NCK-associated protein 1-like |
184 |
0.93 |
| chr11_64215600_64216212 | 0.26 |
AP003774.4 |
HCG1652096, isoform CRA_a; Uncharacterized protein; cDNA FLJ37045 fis, clone BRACE2012185 |
640 |
0.68 |
| chr22_17305454_17306380 | 0.25 |
HSFY1P1 |
heat shock transcription factor, Y-linked 1 pseudogene 1 |
2524 |
0.24 |
| chr2_103034578_103034791 | 0.24 |
IL18RAP |
interleukin 18 receptor accessory protein |
465 |
0.76 |
| chrX_47489873_47490024 | 0.23 |
CFP |
complement factor properdin |
244 |
0.88 |
| chr21_36881240_36881456 | 0.23 |
ENSG00000211590 |
. |
211665 |
0.02 |
| chr22_50242150_50242826 | 0.22 |
ZBED4 |
zinc finger, BED-type containing 4 |
5002 |
0.2 |
| chr12_49616622_49616880 | 0.22 |
TUBA1C |
tubulin, alpha 1c |
4958 |
0.14 |
| chr8_145086794_145086945 | 0.22 |
PARP10 |
poly (ADP-ribose) polymerase family, member 10 |
71 |
0.74 |
| chr10_135091510_135091693 | 0.21 |
ADAM8 |
ADAM metallopeptidase domain 8 |
1229 |
0.29 |
| chr10_111783224_111783375 | 0.21 |
ADD3 |
adducin 3 (gamma) |
15577 |
0.2 |
| chr15_94866308_94866459 | 0.21 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
24888 |
0.28 |
| chr6_10598498_10598818 | 0.20 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
12679 |
0.17 |
| chr15_85315766_85316058 | 0.20 |
ZNF592 |
zinc finger protein 592 |
9973 |
0.14 |
| chr9_93558631_93558871 | 0.19 |
SYK |
spleen tyrosine kinase |
5318 |
0.35 |
| chr8_90789610_90789761 | 0.19 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
19623 |
0.27 |
| chr11_9271619_9271993 | 0.19 |
RP11-5L12.1 |
|
4750 |
0.21 |
| chr14_23218066_23218219 | 0.19 |
ENSG00000212302 |
. |
7896 |
0.12 |
| chr4_139219992_139220143 | 0.18 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
56564 |
0.17 |
| chr12_95043269_95044318 | 0.18 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
545 |
0.85 |
| chr6_114178016_114178613 | 0.18 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
227 |
0.93 |
| chr1_211780463_211780858 | 0.18 |
RP11-359E8.5 |
|
23593 |
0.15 |
| chr5_39177582_39177762 | 0.18 |
FYB |
FYN binding protein |
25457 |
0.23 |
| chr18_2653530_2653681 | 0.18 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
2132 |
0.25 |
| chrX_44203680_44203969 | 0.18 |
EFHC2 |
EF-hand domain (C-terminal) containing 2 |
906 |
0.71 |
| chr2_135070643_135070794 | 0.18 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
58888 |
0.15 |
| chr8_131004829_131004980 | 0.17 |
ENSG00000253043 |
. |
11943 |
0.17 |
| chr9_135855020_135855171 | 0.17 |
GFI1B |
growth factor independent 1B transcription repressor |
997 |
0.42 |
| chr3_186250449_186250600 | 0.17 |
CRYGS |
crystallin, gamma S |
11716 |
0.17 |
| chr1_161578680_161578831 | 0.17 |
FCGR2C |
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene) |
13412 |
0.11 |
| chr17_29639732_29640055 | 0.17 |
EVI2B |
ecotropic viral integration site 2B |
1209 |
0.36 |
| chr16_81852082_81852236 | 0.17 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
39296 |
0.2 |
| chr15_101783225_101783409 | 0.17 |
CHSY1 |
chondroitin sulfate synthase 1 |
8820 |
0.18 |
| chr5_55711467_55711618 | 0.17 |
ENSG00000265665 |
. |
41423 |
0.16 |
| chr2_143829142_143829293 | 0.16 |
ARHGAP15 |
Rho GTPase activating protein 15 |
19714 |
0.25 |
| chr1_110429914_110430065 | 0.16 |
RP11-195M16.1 |
|
1103 |
0.5 |
| chr4_6940579_6940730 | 0.16 |
ENSG00000265953 |
. |
14362 |
0.15 |
| chr19_36444792_36444943 | 0.16 |
LRFN3 |
leucine rich repeat and fibronectin type III domain containing 3 |
16845 |
0.09 |
| chr5_70316349_70316527 | 0.16 |
NAIP |
NLR family, apoptosis inhibitory protein |
99 |
0.98 |
| chr5_17255187_17255338 | 0.16 |
ENSG00000252908 |
. |
14542 |
0.19 |
| chr5_68902997_68903156 | 0.16 |
RP11-848G14.2 |
|
11064 |
0.18 |
| chr1_153363978_153364129 | 0.16 |
S100A8 |
S100 calcium binding protein A8 |
389 |
0.76 |
| chr12_10096266_10096511 | 0.16 |
CLEC12A |
C-type lectin domain family 12, member A |
7527 |
0.14 |
| chr19_28271110_28271360 | 0.16 |
AC005758.1 |
|
138616 |
0.05 |
| chr12_116510107_116510258 | 0.16 |
ENSG00000200665 |
. |
10193 |
0.25 |
| chr17_10102219_10102533 | 0.16 |
GAS7 |
growth arrest-specific 7 |
508 |
0.83 |
| chr5_69758153_69758307 | 0.16 |
RP11-497H16.7 |
|
11045 |
0.27 |
| chr12_110517460_110517751 | 0.16 |
C12orf76 |
chromosome 12 open reading frame 76 |
6114 |
0.22 |
| chr13_41752294_41752445 | 0.16 |
ENSG00000238651 |
. |
1854 |
0.3 |
| chr8_61821706_61823076 | 0.16 |
RP11-33I11.2 |
|
100226 |
0.08 |
| chr21_45509829_45509980 | 0.16 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
17267 |
0.16 |
| chr17_57789761_57790061 | 0.15 |
VMP1 |
vacuole membrane protein 1 |
4828 |
0.18 |
| chr14_38548074_38548225 | 0.15 |
CTD-2058B24.2 |
|
12214 |
0.26 |
| chr6_109699129_109699388 | 0.15 |
CD164 |
CD164 molecule, sialomucin |
3645 |
0.18 |
| chr7_134846065_134846216 | 0.15 |
RP11-134L10.1 |
|
7015 |
0.14 |
| chr6_6687519_6687926 | 0.15 |
LY86-AS1 |
LY86 antisense RNA 1 |
64718 |
0.14 |
| chr12_10019991_10020525 | 0.15 |
RP11-290C10.1 |
|
632 |
0.61 |
| chr19_54345711_54345913 | 0.15 |
NLRP12 |
NLR family, pyrin domain containing 12 |
18164 |
0.05 |
| chr13_49478658_49478809 | 0.15 |
ENSG00000265585 |
. |
25615 |
0.21 |
| chr1_192905339_192905490 | 0.15 |
ENSG00000223075 |
. |
60299 |
0.12 |
| chr4_141015619_141015856 | 0.15 |
RP11-392B6.1 |
|
33432 |
0.2 |
| chr10_131555449_131556065 | 0.15 |
RP11-109A6.2 |
|
55936 |
0.14 |
| chr7_33089731_33090334 | 0.15 |
ENSG00000241420 |
. |
4330 |
0.18 |
| chr1_160673796_160673952 | 0.14 |
CD48 |
CD48 molecule |
7719 |
0.16 |
| chr1_235115156_235115307 | 0.14 |
ENSG00000239690 |
. |
75298 |
0.1 |
| chr2_121354385_121354536 | 0.14 |
ENSG00000201006 |
. |
54389 |
0.16 |
| chr7_2570576_2570806 | 0.14 |
ENSG00000264357 |
. |
3983 |
0.17 |
| chr1_221204797_221204948 | 0.14 |
HLX |
H2.0-like homeobox |
150288 |
0.04 |
| chr11_61738714_61739353 | 0.14 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
3580 |
0.16 |
| chr2_135051726_135051877 | 0.14 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
39971 |
0.19 |
| chrX_20132855_20133006 | 0.14 |
MAP7D2 |
MAP7 domain containing 2 |
1820 |
0.28 |
| chr6_64062151_64062302 | 0.14 |
LGSN |
lengsin, lens protein with glutamine synthetase domain |
32344 |
0.24 |
| chr6_24932242_24932425 | 0.14 |
FAM65B |
family with sequence similarity 65, member B |
3855 |
0.28 |
| chr5_14560333_14560610 | 0.14 |
FAM105A |
family with sequence similarity 105, member A |
21413 |
0.23 |
| chr20_44689695_44689846 | 0.14 |
NCOA5 |
nuclear receptor coactivator 5 |
28821 |
0.11 |
| chr3_13002263_13002414 | 0.14 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
6830 |
0.26 |
| chr7_112062054_112062259 | 0.14 |
IFRD1 |
interferon-related developmental regulator 1 |
867 |
0.64 |
| chr4_55534639_55534790 | 0.13 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
10629 |
0.31 |
| chr4_141174038_141174780 | 0.13 |
SCOC |
short coiled-coil protein |
4031 |
0.26 |
| chr9_84494361_84494512 | 0.13 |
ENSG00000252167 |
. |
5975 |
0.17 |
| chr3_63793614_63793765 | 0.13 |
AC136289.1 |
|
2920 |
0.24 |
| chrX_135961684_135962842 | 0.13 |
RBMX |
RNA binding motif protein, X-linked |
621 |
0.53 |
| chr20_39657787_39658990 | 0.13 |
TOP1 |
topoisomerase (DNA) I |
930 |
0.61 |
| chr1_16160235_16160851 | 0.13 |
RP11-169K16.9 |
|
1812 |
0.28 |
| chr17_73774322_73775453 | 0.13 |
H3F3B |
H3 histone, family 3B (H3.3B) |
480 |
0.64 |
| chr5_134761710_134761905 | 0.13 |
C5orf20 |
chromosome 5 open reading frame 20 |
21231 |
0.12 |
| chr7_140615501_140615652 | 0.13 |
BRAF |
v-raf murine sarcoma viral oncogene homolog B |
8988 |
0.22 |
| chr10_3527831_3528010 | 0.13 |
RP11-184A2.3 |
|
265339 |
0.02 |
| chr1_3527588_3528047 | 0.13 |
MEGF6 |
multiple EGF-like-domains 6 |
242 |
0.89 |
| chr1_204940483_204940634 | 0.13 |
NFASC |
neurofascin |
10426 |
0.21 |
| chr6_105153386_105153605 | 0.13 |
ENSG00000252944 |
. |
61202 |
0.13 |
| chr9_297994_298145 | 0.13 |
DOCK8 |
dedicator of cytokinesis 8 |
24999 |
0.18 |
| chr15_93426669_93427240 | 0.12 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
428 |
0.83 |
| chr20_48887078_48887229 | 0.12 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
79777 |
0.09 |
| chr7_105998110_105998261 | 0.12 |
NAMPT |
nicotinamide phosphoribosyltransferase |
71413 |
0.11 |
| chr17_66433856_66434617 | 0.12 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
4524 |
0.23 |
| chr2_68953487_68953638 | 0.12 |
ARHGAP25 |
Rho GTPase activating protein 25 |
8351 |
0.26 |
| chrX_135738768_135738919 | 0.12 |
CD40LG |
CD40 ligand |
8457 |
0.18 |
| chr12_93163278_93163429 | 0.12 |
PLEKHG7 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 7 |
33042 |
0.18 |
| chr1_161600101_161600252 | 0.12 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
646 |
0.42 |
| chr3_46970853_46971178 | 0.12 |
PTH1R |
parathyroid hormone 1 receptor |
26926 |
0.13 |
| chr14_24641513_24641917 | 0.12 |
REC8 |
REC8 meiotic recombination protein |
105 |
0.89 |
| chr11_65378670_65378821 | 0.12 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
27 |
0.93 |
| chr12_57870819_57871184 | 0.12 |
ARHGAP9 |
Rho GTPase activating protein 9 |
566 |
0.53 |
| chr22_36632572_36632723 | 0.12 |
APOL2 |
apolipoprotein L, 2 |
2626 |
0.25 |
| chr12_121676925_121677077 | 0.12 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
21344 |
0.16 |
| chr3_184395596_184395747 | 0.12 |
MAGEF1 |
melanoma antigen family F, 1 |
34165 |
0.19 |
| chr3_36937026_36937306 | 0.12 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
36792 |
0.17 |
| chr2_169940004_169940155 | 0.12 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
2462 |
0.31 |
| chr13_31943501_31943652 | 0.12 |
B3GALTL |
beta 1,3-galactosyltransferase-like |
169503 |
0.04 |
| chr4_153600093_153601094 | 0.12 |
TMEM154 |
transmembrane protein 154 |
571 |
0.78 |
| chr7_104584020_104584171 | 0.12 |
ENSG00000251911 |
. |
27639 |
0.16 |
| chr1_150550029_150550609 | 0.11 |
MCL1 |
myeloid cell leukemia sequence 1 (BCL2-related) |
1687 |
0.18 |
| chr3_152939222_152939486 | 0.11 |
ENSG00000265813 |
. |
21548 |
0.22 |
| chr18_9701912_9702261 | 0.11 |
RAB31 |
RAB31, member RAS oncogene family |
6076 |
0.24 |
| chr18_2960483_2961188 | 0.11 |
RP11-737O24.1 |
|
6181 |
0.16 |
| chr5_77807061_77807212 | 0.11 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
37838 |
0.21 |
| chr12_80321477_80321628 | 0.11 |
PPP1R12A |
protein phosphatase 1, regulatory subunit 12A |
6943 |
0.2 |
| chr13_77324135_77324286 | 0.11 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
136315 |
0.05 |
| chr5_37317955_37318106 | 0.11 |
ENSG00000251880 |
. |
9207 |
0.22 |
| chr6_39282333_39282484 | 0.11 |
KCNK17 |
potassium channel, subfamily K, member 17 |
79 |
0.98 |
| chr17_27256722_27256873 | 0.11 |
PHF12 |
PHD finger protein 12 |
1489 |
0.19 |
| chr6_89754695_89754846 | 0.11 |
ENSG00000223001 |
. |
18639 |
0.15 |
| chr11_82608630_82608781 | 0.11 |
C11orf82 |
chromosome 11 open reading frame 82 |
2312 |
0.28 |
| chr11_48088687_48088966 | 0.11 |
ENSG00000263693 |
. |
29508 |
0.17 |
| chr7_5710794_5710945 | 0.11 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
9223 |
0.21 |
| chr4_88896230_88897032 | 0.11 |
SPP1 |
secreted phosphoprotein 1 |
188 |
0.95 |
| chr2_223536011_223536232 | 0.11 |
MOGAT1 |
monoacylglycerol O-acyltransferase 1 |
384 |
0.86 |
| chr10_81947502_81948014 | 0.11 |
ANXA11 |
annexin A11 |
14981 |
0.2 |
| chr19_16482667_16482930 | 0.11 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
10034 |
0.15 |
| chr18_379614_379765 | 0.11 |
RP11-720L2.2 |
|
44727 |
0.16 |
| chr15_99074007_99074158 | 0.11 |
FAM169B |
family with sequence similarity 169, member B |
16471 |
0.27 |
| chr2_192604587_192604738 | 0.11 |
AC098872.3 |
|
53489 |
0.14 |
| chr5_119593776_119593927 | 0.11 |
ENSG00000251975 |
. |
79497 |
0.12 |
| chr14_64806720_64806871 | 0.11 |
RP11-544I20.2 |
|
1478 |
0.34 |
| chr16_27301591_27301742 | 0.11 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
21551 |
0.15 |
| chr12_805861_806012 | 0.11 |
NINJ2 |
ninjurin 2 |
32991 |
0.15 |
| chr9_7653385_7653536 | 0.11 |
TMEM261 |
transmembrane protein 261 |
146607 |
0.05 |
| chr5_126147731_126147991 | 0.11 |
LMNB1 |
lamin B1 |
34980 |
0.19 |
| chr7_143080537_143080688 | 0.10 |
ZYX |
zyxin |
519 |
0.65 |
| chr1_61543284_61543773 | 0.10 |
NFIA |
nuclear factor I/A |
582 |
0.78 |
| chr2_201987013_201987300 | 0.10 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
44 |
0.96 |
| chr12_110562218_110563151 | 0.10 |
IFT81 |
intraflagellar transport 81 homolog (Chlamydomonas) |
487 |
0.84 |
| chr21_34602307_34603720 | 0.10 |
IFNAR2 |
interferon (alpha, beta and omega) receptor 2 |
299 |
0.87 |
| chr20_8583214_8583783 | 0.10 |
ENSG00000221201 |
. |
6329 |
0.25 |
| chr1_183556161_183556312 | 0.10 |
NCF2 |
neutrophil cytosolic factor 2 |
3480 |
0.25 |
| chr15_92012895_92013046 | 0.10 |
SV2B |
synaptic vesicle glycoprotein 2B |
243870 |
0.02 |
| chr8_70650371_70650522 | 0.10 |
RP11-102F4.2 |
|
24821 |
0.19 |
| chr7_23246593_23246744 | 0.10 |
NUPL2 |
nucleoporin like 2 |
24985 |
0.16 |
| chr6_42537305_42537764 | 0.10 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
5734 |
0.24 |
| chr17_68678856_68679007 | 0.10 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
513255 |
0.0 |
| chr8_90995843_90996298 | 0.10 |
NBN |
nibrin |
395 |
0.87 |
| chr4_6909699_6909850 | 0.10 |
TBC1D14 |
TBC1 domain family, member 14 |
1195 |
0.49 |
| chr20_45028360_45028511 | 0.10 |
ELMO2 |
engulfment and cell motility 2 |
5314 |
0.24 |
| chr11_94335838_94335989 | 0.10 |
PIWIL4 |
piwi-like RNA-mediated gene silencing 4 |
15042 |
0.2 |
| chr8_27141347_27141498 | 0.10 |
TRIM35 |
tripartite motif containing 35 |
10230 |
0.19 |
| chr13_48809687_48810120 | 0.10 |
ITM2B |
integral membrane protein 2B |
2564 |
0.37 |
| chr2_33355355_33355506 | 0.10 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
4043 |
0.34 |
| chr11_93451363_93451537 | 0.10 |
ENSG00000199875 |
. |
1319 |
0.18 |
| chr15_92404035_92404308 | 0.10 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
6820 |
0.27 |
| chr7_155484798_155484949 | 0.10 |
RBM33 |
RNA binding motif protein 33 |
8712 |
0.25 |
| chr6_138176930_138177081 | 0.10 |
RP11-356I2.4 |
|
2180 |
0.35 |
| chr10_102106243_102106464 | 0.10 |
RP11-34D15.2 |
|
206 |
0.76 |
| chr1_164489142_164489293 | 0.10 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
39220 |
0.2 |
| chr1_29241076_29241272 | 0.10 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
83 |
0.97 |
| chr2_218990770_218991030 | 0.10 |
CXCR2 |
chemokine (C-X-C motif) receptor 2 |
158 |
0.95 |
| chr2_48338474_48339359 | 0.10 |
ENSG00000201010 |
. |
105798 |
0.07 |
| chr1_21503574_21503811 | 0.10 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
315 |
0.69 |
| chr4_79583127_79583675 | 0.10 |
ENSG00000238816 |
. |
22147 |
0.19 |
| chr1_207495975_207496182 | 0.10 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
942 |
0.68 |
| chr12_40547331_40547482 | 0.10 |
LRRK2 |
leucine-rich repeat kinase 2 |
43140 |
0.16 |
| chr19_23427286_23427437 | 0.09 |
ZNF724P |
zinc finger protein 724, pseudogene |
5801 |
0.26 |
| chr18_72163609_72164283 | 0.09 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
328 |
0.9 |
| chr21_37557715_37557866 | 0.09 |
DOPEY2 |
dopey family member 2 |
20957 |
0.12 |
| chr1_200335563_200335714 | 0.09 |
ZNF281 |
zinc finger protein 281 |
43482 |
0.2 |
| chr5_98265881_98266063 | 0.09 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
3732 |
0.25 |
| chr5_1501228_1501379 | 0.09 |
LPCAT1 |
lysophosphatidylcholine acyltransferase 1 |
22789 |
0.19 |
| chrX_119737965_119739054 | 0.09 |
MCTS1 |
malignant T cell amplified sequence 1 |
333 |
0.88 |
| chr14_92942291_92942442 | 0.09 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
36658 |
0.18 |
| chr3_119814638_119815091 | 0.09 |
GSK3B |
glycogen synthase kinase 3 beta |
1600 |
0.39 |
| chr4_10642308_10642459 | 0.09 |
CLNK |
cytokine-dependent hematopoietic cell linker |
15802 |
0.3 |
| chr2_70474548_70475674 | 0.09 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
475 |
0.73 |
| chr10_90593785_90593936 | 0.09 |
ANKRD22 |
ankyrin repeat domain 22 |
17715 |
0.16 |
| chr2_143909648_143909799 | 0.09 |
RP11-190J23.1 |
|
20018 |
0.22 |
| chr12_32740907_32741058 | 0.09 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
10478 |
0.26 |
| chr9_108104857_108105008 | 0.09 |
SLC44A1 |
solute carrier family 44 (choline transporter), member 1 |
12825 |
0.28 |
| chr16_17547681_17547832 | 0.09 |
XYLT1 |
xylosyltransferase I |
16982 |
0.31 |
| chr16_30103433_30103902 | 0.09 |
TBX6 |
T-box 6 |
459 |
0.59 |
| chr8_38325453_38326142 | 0.09 |
FGFR1 |
fibroblast growth factor receptor 1 |
337 |
0.87 |
| chr10_49922246_49922397 | 0.09 |
WDFY4 |
WDFY family member 4 |
4570 |
0.28 |
| chr1_84610970_84611403 | 0.09 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
1232 |
0.6 |
| chr6_53130180_53130331 | 0.09 |
ENSG00000264056 |
. |
11536 |
0.19 |
| chr9_117721707_117721858 | 0.09 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
29085 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.1 | 0.2 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:2000380 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0042663 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.0 | GO:0000239 | pachytene(GO:0000239) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:1904019 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.0 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.0 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.3 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0004339 | glucan 1,4-alpha-glucosidase activity(GO:0004339) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.0 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.0 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.3 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |