Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
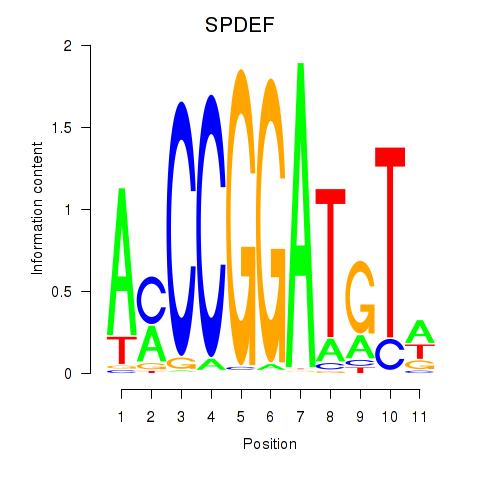
Results for SPDEF
Z-value: 0.25
Transcription factors associated with SPDEF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SPDEF
|
ENSG00000124664.6 | SAM pointed domain containing ETS transcription factor |
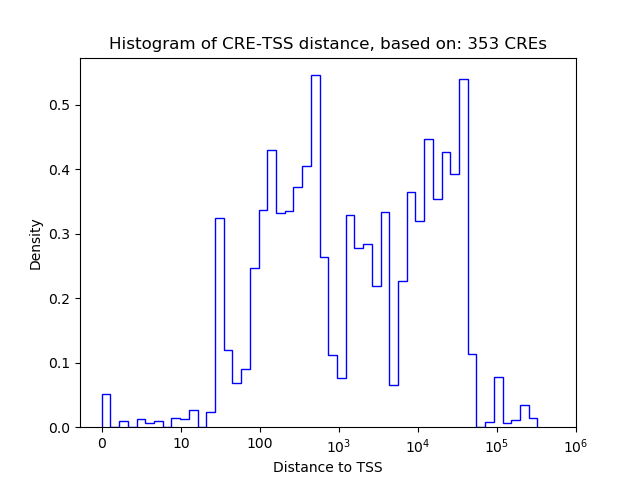
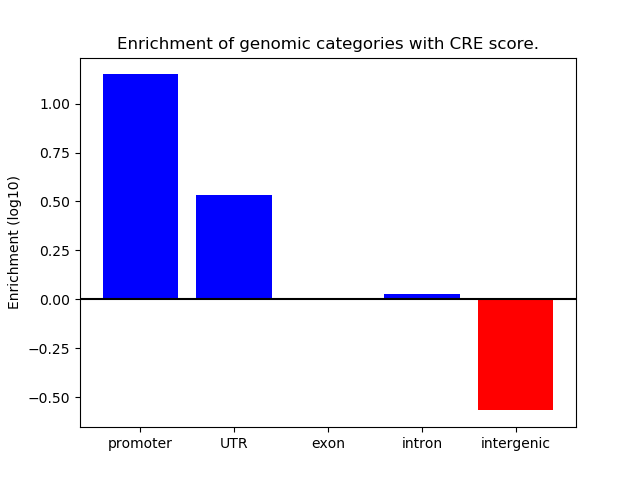
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_34544180_34544427 | SPDEF | 20193 | 0.175331 | -0.26 | 5.0e-01 | Click! |
Activity of the SPDEF motif across conditions
Conditions sorted by the z-value of the SPDEF motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
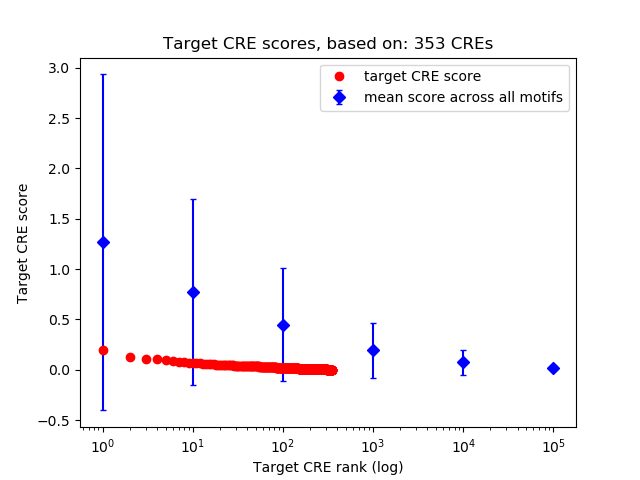
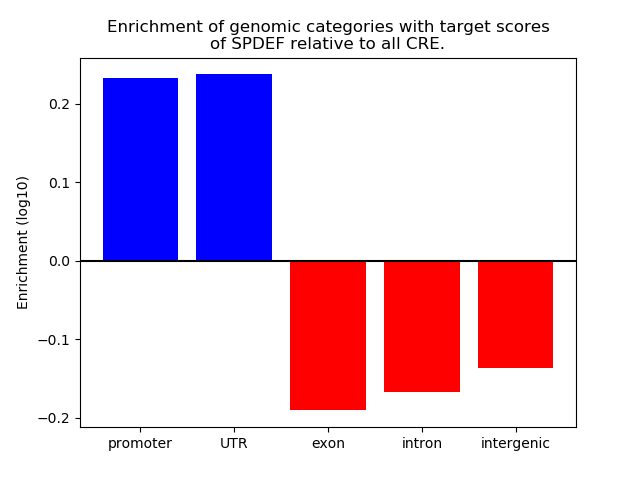
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_160616915_160617313 | 0.20 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
29 |
0.97 |
| chr14_25143618_25143855 | 0.13 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
40263 |
0.14 |
| chr17_76129934_76130268 | 0.11 |
TMC6 |
transmembrane channel-like 6 |
1613 |
0.24 |
| chr4_38806937_38807296 | 0.10 |
TLR1 |
toll-like receptor 1 |
83 |
0.97 |
| chr15_101061059_101061210 | 0.10 |
CERS3 |
ceramide synthase 3 |
23342 |
0.14 |
| chr19_18287388_18287660 | 0.09 |
IFI30 |
interferon, gamma-inducible protein 30 |
3047 |
0.14 |
| chr10_30994119_30994515 | 0.08 |
SVILP1 |
supervillin pseudogene 1 |
9467 |
0.26 |
| chr16_27378896_27379134 | 0.07 |
IL4R |
interleukin 4 receptor |
15019 |
0.19 |
| chr12_133052078_133052229 | 0.07 |
MUC8 |
mucin 8 |
1427 |
0.44 |
| chr11_119600715_119600866 | 0.07 |
CTD-2523D13.2 |
|
497 |
0.68 |
| chr17_45356556_45356707 | 0.07 |
ENSG00000238419 |
. |
19845 |
0.12 |
| chr19_35949823_35949974 | 0.07 |
FFAR2 |
free fatty acid receptor 2 |
9281 |
0.11 |
| chr12_53496670_53496821 | 0.06 |
SOAT2 |
sterol O-acyltransferase 2 |
557 |
0.63 |
| chr16_11654733_11655066 | 0.06 |
LITAF |
lipopolysaccharide-induced TNF factor |
25330 |
0.15 |
| chr14_24615202_24615772 | 0.05 |
PSME2 |
proteasome (prosome, macropain) activator subunit 2 (PA28 beta) |
36 |
0.82 |
| chr18_43266884_43267229 | 0.05 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
20949 |
0.17 |
| chr21_16424536_16424754 | 0.05 |
NRIP1 |
nuclear receptor interacting protein 1 |
12481 |
0.26 |
| chrX_134429275_134429737 | 0.05 |
ZNF75D |
zinc finger protein 75D |
453 |
0.86 |
| chr6_31794829_31795180 | 0.05 |
HSPA1B |
heat shock 70kDa protein 1B |
508 |
0.47 |
| chr21_40734232_40734404 | 0.05 |
HMGN1 |
high mobility group nucleosome binding domain 1 |
12745 |
0.12 |
| chr20_62198919_62199070 | 0.05 |
HELZ2 |
helicase with zinc finger 2, transcriptional coactivator |
433 |
0.7 |
| chr20_25038810_25039334 | 0.05 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
254 |
0.93 |
| chr8_1954691_1954842 | 0.05 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
32722 |
0.19 |
| chrX_13161411_13161562 | 0.05 |
FAM9C |
family with sequence similarity 9, member C |
98685 |
0.08 |
| chr6_35279100_35279566 | 0.05 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
1818 |
0.34 |
| chr2_99349001_99349152 | 0.05 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
1487 |
0.47 |
| chr19_10676975_10677686 | 0.04 |
KRI1 |
KRI1 homolog (S. cerevisiae) |
617 |
0.51 |
| chr5_60922081_60922360 | 0.04 |
C5orf64 |
chromosome 5 open reading frame 64 |
11416 |
0.22 |
| chr7_156811762_156812002 | 0.04 |
MNX1-AS1 |
MNX1 antisense RNA 1 (head to head) |
8383 |
0.17 |
| chr1_36042987_36043882 | 0.04 |
RP4-728D4.2 |
|
104 |
0.95 |
| chr17_4934347_4934505 | 0.04 |
SLC52A1 |
solute carrier family 52 (riboflavin transporter), member 1 |
4295 |
0.09 |
| chr19_7985929_7986556 | 0.04 |
SNAPC2 |
small nuclear RNA activating complex, polypeptide 2, 45kDa |
342 |
0.66 |
| chr17_72744920_72745438 | 0.04 |
MIR3615 |
microRNA 3615 |
128 |
0.67 |
| chr16_31880184_31880698 | 0.04 |
ZNF267 |
zinc finger protein 267 |
4638 |
0.26 |
| chr12_11923358_11923509 | 0.04 |
ETV6 |
ets variant 6 |
17998 |
0.27 |
| chr2_220112643_220112927 | 0.04 |
STK16 |
serine/threonine kinase 16 |
2167 |
0.11 |
| chr12_40622573_40622724 | 0.04 |
LRRK2 |
leucine-rich repeat kinase 2 |
3772 |
0.27 |
| chr17_41438253_41439076 | 0.04 |
ENSG00000188825 |
. |
26121 |
0.1 |
| chr15_75082239_75082553 | 0.04 |
ENSG00000264386 |
. |
1298 |
0.27 |
| chr16_8738665_8738816 | 0.04 |
METTL22 |
methyltransferase like 22 |
2415 |
0.28 |
| chr9_126101731_126102049 | 0.04 |
CRB2 |
crumbs homolog 2 (Drosophila) |
16559 |
0.22 |
| chr5_142185385_142185536 | 0.04 |
ARHGAP26 |
Rho GTPase activating protein 26 |
33178 |
0.2 |
| chrX_62780920_62781228 | 0.04 |
ENSG00000235437 |
. |
123 |
0.98 |
| chr6_24936325_24936476 | 0.04 |
FAM65B |
family with sequence similarity 65, member B |
212 |
0.95 |
| chr8_131218803_131218954 | 0.04 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
25770 |
0.21 |
| chr6_155504502_155504653 | 0.04 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
33194 |
0.19 |
| chr2_201991504_201992128 | 0.04 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
2236 |
0.22 |
| chr18_53256970_53257121 | 0.04 |
TCF4 |
transcription factor 4 |
0 |
0.99 |
| chr2_220083227_220083846 | 0.03 |
ABCB6 |
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
136 |
0.88 |
| chr19_42052040_42052310 | 0.03 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
3711 |
0.2 |
| chr14_105864635_105864945 | 0.03 |
TEX22 |
testis expressed 22 |
130 |
0.94 |
| chr2_238604951_238605166 | 0.03 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
4076 |
0.27 |
| chr3_53284311_53284462 | 0.03 |
TKT |
transketolase |
5630 |
0.16 |
| chr3_12705860_12706283 | 0.03 |
RAF1 |
v-raf-1 murine leukemia viral oncogene homolog 1 |
346 |
0.88 |
| chr1_9295235_9295615 | 0.03 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
591 |
0.76 |
| chr11_71639675_71639978 | 0.03 |
RNF121 |
ring finger protein 121 |
79 |
0.56 |
| chr5_126147731_126147991 | 0.03 |
LMNB1 |
lamin B1 |
34980 |
0.19 |
| chr1_208056352_208056503 | 0.03 |
CD34 |
CD34 molecule |
7410 |
0.29 |
| chr10_61632198_61632349 | 0.03 |
CCDC6 |
coiled-coil domain containing 6 |
34141 |
0.21 |
| chr11_108108232_108108383 | 0.03 |
ENSG00000206967 |
. |
7976 |
0.16 |
| chr2_240302618_240303103 | 0.03 |
HDAC4 |
histone deacetylase 4 |
19783 |
0.17 |
| chr15_68121116_68121316 | 0.03 |
SKOR1 |
SKI family transcriptional corepressor 1 |
3275 |
0.3 |
| chr2_86088659_86088948 | 0.03 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
5974 |
0.2 |
| chr16_30759296_30759478 | 0.03 |
PHKG2 |
phosphorylase kinase, gamma 2 (testis) |
204 |
0.86 |
| chr1_208041477_208041725 | 0.03 |
CD34 |
CD34 molecule |
22236 |
0.23 |
| chr14_92942291_92942442 | 0.03 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
36658 |
0.18 |
| chr11_44586532_44586926 | 0.03 |
CD82 |
CD82 molecule |
412 |
0.88 |
| chr1_40204287_40204449 | 0.03 |
PPIE |
peptidylprolyl isomerase E (cyclophilin E) |
186 |
0.92 |
| chr1_42174997_42175148 | 0.03 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
8400 |
0.27 |
| chr17_19015939_19016273 | 0.03 |
ENSG00000262202 |
. |
157 |
0.9 |
| chr17_18964908_18965219 | 0.03 |
ENSG00000265185 |
. |
162 |
0.91 |
| chr17_19091011_19091355 | 0.03 |
ENSG00000263934 |
. |
146 |
0.9 |
| chr17_19093552_19093880 | 0.03 |
ENSG00000264940 |
. |
158 |
0.9 |
| chr17_18967446_18967725 | 0.03 |
ENSG00000262074 |
. |
136 |
0.92 |
| chr13_96130880_96131359 | 0.03 |
CLDN10 |
claudin 10 |
45261 |
0.16 |
| chr6_33168747_33168964 | 0.03 |
SLC39A7 |
solute carrier family 39 (zinc transporter), member 7 |
205 |
0.48 |
| chr16_53468750_53469435 | 0.03 |
RBL2 |
retinoblastoma-like 2 (p130) |
439 |
0.8 |
| chrX_146992887_146993246 | 0.03 |
ENSG00000268066 |
. |
269 |
0.62 |
| chrX_53712878_53713294 | 0.03 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
587 |
0.83 |
| chr11_71751582_71752578 | 0.02 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
103 |
0.93 |
| chr5_77147504_77148481 | 0.02 |
TBCA |
tubulin folding cofactor A |
16612 |
0.28 |
| chr7_142430227_142430392 | 0.02 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
27010 |
0.17 |
| chr14_31028402_31029308 | 0.02 |
G2E3 |
G2/M-phase specific E3 ubiquitin protein ligase |
339 |
0.9 |
| chr19_10831379_10831550 | 0.02 |
DNM2 |
dynamin 2 |
2227 |
0.18 |
| chr8_19462920_19463071 | 0.02 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
2989 |
0.41 |
| chr13_51490802_51491019 | 0.02 |
RNASEH2B-AS1 |
RNASEH2B antisense RNA 1 |
6062 |
0.21 |
| chr1_155202832_155202983 | 0.02 |
ENSG00000216109 |
. |
3721 |
0.07 |
| chr20_52239219_52239370 | 0.02 |
ZNF217 |
zinc finger protein 217 |
13863 |
0.2 |
| chr5_126323849_126324159 | 0.02 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
42496 |
0.18 |
| chr1_8455141_8455292 | 0.02 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
28510 |
0.15 |
| chr5_176852709_176853043 | 0.02 |
GRK6 |
G protein-coupled receptor kinase 6 |
621 |
0.54 |
| chr3_128722489_128722640 | 0.02 |
EFCC1 |
EF-hand and coiled-coil domain containing 1 |
2092 |
0.25 |
| chr4_84309529_84309680 | 0.02 |
HPSE |
heparanase |
53298 |
0.12 |
| chr1_114474532_114474797 | 0.02 |
HIPK1 |
homeodomain interacting protein kinase 1 |
1314 |
0.32 |
| chr1_65533362_65534513 | 0.02 |
ENSG00000199135 |
. |
9746 |
0.19 |
| chr6_44027253_44027404 | 0.02 |
RP5-1120P11.1 |
|
15061 |
0.17 |
| chr13_26796516_26796828 | 0.02 |
RNF6 |
ring finger protein (C3H2C3 type) 6 |
119 |
0.98 |
| chr14_61966690_61966841 | 0.02 |
RP11-47I22.4 |
|
29081 |
0.17 |
| chr7_99097527_99097880 | 0.02 |
ZNF394 |
zinc finger protein 394 |
184 |
0.89 |
| chr2_69009313_69009464 | 0.02 |
ARHGAP25 |
Rho GTPase activating protein 25 |
7316 |
0.24 |
| chr14_64969597_64970324 | 0.02 |
ZBTB1 |
zinc finger and BTB domain containing 1 |
483 |
0.49 |
| chr21_45779191_45779342 | 0.02 |
TRPM2 |
transient receptor potential cation channel, subfamily M, member 2 |
5695 |
0.1 |
| chr17_57231407_57232561 | 0.02 |
SKA2 |
spindle and kinetochore associated complex subunit 2 |
551 |
0.48 |
| chr11_46143390_46143562 | 0.02 |
PHF21A |
PHD finger protein 21A |
491 |
0.82 |
| chr3_11683730_11684257 | 0.02 |
VGLL4 |
vestigial like 4 (Drosophila) |
1405 |
0.47 |
| chrY_21728540_21728933 | 0.02 |
TXLNG2P |
taxilin gamma 2, pseudogene |
499 |
0.84 |
| chr3_50388284_50388569 | 0.02 |
NPRL2 |
nitrogen permease regulator-like 2 (S. cerevisiae) |
96 |
0.35 |
| chr12_123921440_123921638 | 0.02 |
RILPL2 |
Rab interacting lysosomal protein-like 2 |
275 |
0.89 |
| chr16_57168008_57168159 | 0.02 |
CPNE2 |
copine II |
14972 |
0.14 |
| chr5_142164826_142164977 | 0.02 |
ARHGAP26 |
Rho GTPase activating protein 26 |
12619 |
0.26 |
| chr14_22950797_22950948 | 0.02 |
TRAJ55 |
T cell receptor alpha joining 55 (pseudogene) |
186 |
0.79 |
| chr4_154354394_154354673 | 0.02 |
KIAA0922 |
KIAA0922 |
32965 |
0.17 |
| chr11_576246_576634 | 0.02 |
PHRF1 |
PHD and ring finger domains 1 |
46 |
0.93 |
| chr8_10872806_10873547 | 0.02 |
XKR6 |
XK, Kell blood group complex subunit-related family, member 6 |
13185 |
0.18 |
| chr20_44562997_44563218 | 0.02 |
PCIF1 |
PDX1 C-terminal inhibiting factor 1 |
160 |
0.91 |
| chr20_48531824_48532295 | 0.02 |
SPATA2 |
spermatogenesis associated 2 |
14 |
0.97 |
| chr12_89199825_89199976 | 0.02 |
ENSG00000252850 |
. |
224484 |
0.02 |
| chr7_6098442_6098863 | 0.02 |
EIF2AK1 |
eukaryotic translation initiation factor 2-alpha kinase 1 |
143 |
0.9 |
| chr1_101361069_101361411 | 0.02 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
314 |
0.5 |
| chr2_38323518_38323669 | 0.02 |
CYP1B1-AS1 |
CYP1B1 antisense RNA 1 |
2129 |
0.31 |
| chr7_132766918_132767069 | 0.02 |
CHCHD3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
151 |
0.87 |
| chr19_41082761_41083290 | 0.02 |
SHKBP1 |
SH3KBP1 binding protein 1 |
39 |
0.97 |
| chr11_1595063_1595214 | 0.02 |
KRTAP5-AS1 |
KRTAP5-1/KRTAP5-2 antisense RNA 1 |
259 |
0.85 |
| chr6_139309569_139309822 | 0.02 |
REPS1 |
RALBP1 associated Eps domain containing 1 |
297 |
0.93 |
| chrX_2664260_2664411 | 0.02 |
XG |
Xg blood group |
5756 |
0.23 |
| chr5_56205884_56206600 | 0.02 |
AC008937.3 |
|
299 |
0.48 |
| chr1_9686401_9686552 | 0.02 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
25314 |
0.14 |
| chr19_2888884_2889187 | 0.02 |
ZNF57 |
zinc finger protein 57 |
11861 |
0.1 |
| chr10_12256838_12257395 | 0.01 |
CDC123 |
cell division cycle 123 |
15831 |
0.13 |
| chr7_99647395_99647894 | 0.01 |
ZSCAN21 |
zinc finger and SCAN domain containing 21 |
227 |
0.84 |
| chr4_100868040_100868504 | 0.01 |
DNAJB14 |
DnaJ (Hsp40) homolog, subfamily B, member 14 |
389 |
0.82 |
| chr15_101389926_101390355 | 0.01 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
27779 |
0.19 |
| chr9_95527351_95527701 | 0.01 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
432 |
0.84 |
| chrX_154298671_154299361 | 0.01 |
MTCP1 |
mature T-cell proliferation 1 |
485 |
0.41 |
| chr8_61495922_61496073 | 0.01 |
RAB2A |
RAB2A, member RAS oncogene family |
1285 |
0.53 |
| chr11_77850351_77850689 | 0.01 |
ALG8 |
ALG8, alpha-1,3-glucosyltransferase |
118 |
0.72 |
| chr16_4524699_4524966 | 0.01 |
NMRAL1 |
NmrA-like family domain containing 1 |
64 |
0.56 |
| chr4_13532847_13532998 | 0.01 |
NKX3-2 |
NK3 homeobox 2 |
13752 |
0.22 |
| chr6_167411857_167412522 | 0.01 |
RP1-167A14.2 |
|
364 |
0.49 |
| chr15_90920057_90920398 | 0.01 |
IQGAP1 |
IQ motif containing GTPase activating protein 1 |
11223 |
0.13 |
| chr1_161102671_161103093 | 0.01 |
DEDD |
death effector domain containing |
404 |
0.63 |
| chr11_124670044_124670876 | 0.01 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
109 |
0.49 |
| chr22_18400717_18400868 | 0.01 |
MICAL3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
11152 |
0.2 |
| chr1_47694555_47694706 | 0.01 |
TAL1 |
T-cell acute lymphocytic leukemia 1 |
813 |
0.55 |
| chr6_33172124_33172275 | 0.01 |
HSD17B8 |
hydroxysteroid (17-beta) dehydrogenase 8 |
220 |
0.82 |
| chr12_6472631_6473046 | 0.01 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
475 |
0.7 |
| chr3_108308362_108309163 | 0.01 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
140 |
0.82 |
| chr17_75448601_75448829 | 0.01 |
SEPT9 |
septin 9 |
1192 |
0.41 |
| chr17_35968260_35968971 | 0.01 |
SYNRG |
synergin, gamma |
814 |
0.47 |
| chr14_90848618_90849160 | 0.01 |
ENSG00000238758 |
. |
1347 |
0.42 |
| chr3_194277134_194277288 | 0.01 |
ENSG00000252053 |
. |
22899 |
0.12 |
| chr12_7046177_7046619 | 0.01 |
C12orf57 |
chromosome 12 open reading frame 57 |
6267 |
0.06 |
| chr11_46581907_46582403 | 0.01 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
30759 |
0.15 |
| chr8_82059369_82059520 | 0.01 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
35141 |
0.21 |
| chr17_37844122_37844360 | 0.01 |
PGAP3 |
post-GPI attachment to proteins 3 |
69 |
0.49 |
| chr17_41323238_41323851 | 0.01 |
NBR1 |
neighbor of BRCA1 gene 1 |
284 |
0.87 |
| chrX_152200749_152200932 | 0.01 |
PNMA3 |
paraneoplastic Ma antigen 3 |
23926 |
0.12 |
| chr14_68086133_68086429 | 0.01 |
ARG2 |
arginase 2 |
234 |
0.87 |
| chr19_54714818_54714969 | 0.01 |
RPS9 |
ribosomal protein S9 |
9865 |
0.07 |
| chr1_202976092_202976359 | 0.01 |
TMEM183A |
transmembrane protein 183A |
289 |
0.86 |
| chr16_85608858_85609009 | 0.01 |
GSE1 |
Gse1 coiled-coil protein |
36082 |
0.17 |
| chr19_11200787_11200965 | 0.01 |
LDLR |
low density lipoprotein receptor |
399 |
0.82 |
| chr18_32956679_32957498 | 0.01 |
ZNF396 |
zinc finger protein 396 |
186 |
0.96 |
| chr22_22337183_22337444 | 0.01 |
TOP3B |
topoisomerase (DNA) III beta |
100 |
0.94 |
| chr8_100025267_100025604 | 0.01 |
VPS13B |
vacuolar protein sorting 13 homolog B (yeast) |
59 |
0.97 |
| chr15_89949662_89949989 | 0.01 |
ENSG00000207819 |
. |
38577 |
0.14 |
| chr2_27805637_27805914 | 0.01 |
ZNF512 |
zinc finger protein 512 |
122 |
0.9 |
| chr3_190331805_190331956 | 0.01 |
IL1RAP |
interleukin 1 receptor accessory protein |
1217 |
0.57 |
| chr6_111198520_111198699 | 0.01 |
AMD1 |
adenosylmethionine decarboxylase 1 |
1831 |
0.3 |
| chr2_74682121_74682812 | 0.01 |
INO80B |
INO80 complex subunit B |
223 |
0.81 |
| chr10_65280480_65280949 | 0.01 |
REEP3 |
receptor accessory protein 3 |
409 |
0.89 |
| chr4_1161098_1161472 | 0.01 |
RP11-20I20.4 |
|
281 |
0.86 |
| chr5_154140674_154140825 | 0.01 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
4015 |
0.22 |
| chrX_128978019_128978341 | 0.01 |
ZDHHC9 |
zinc finger, DHHC-type containing 9 |
295 |
0.91 |
| chr12_90103618_90104485 | 0.01 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
974 |
0.58 |
| chr20_23330489_23330738 | 0.01 |
NXT1 |
NTF2-like export factor 1 |
760 |
0.44 |
| chr17_80255580_80256204 | 0.01 |
CD7 |
CD7 molecule |
19536 |
0.1 |
| chr8_97127078_97127229 | 0.01 |
GDF6 |
growth differentiation factor 6 |
45867 |
0.17 |
| chrX_39949292_39950247 | 0.01 |
BCOR |
BCL6 corepressor |
6887 |
0.33 |
| chr2_234263629_234264304 | 0.01 |
DGKD |
diacylglycerol kinase, delta 130kDa |
812 |
0.51 |
| chr13_52586601_52587118 | 0.01 |
ALG11 |
ALG11, alpha-1,2-mannosyltransferase |
309 |
0.84 |
| chr12_113623531_113623954 | 0.01 |
RITA1 |
RBPJ interacting and tubulin associated 1 |
395 |
0.48 |
| chr2_67624524_67625302 | 0.01 |
ETAA1 |
Ewing tumor-associated antigen 1 |
462 |
0.91 |
| chr17_66243472_66244013 | 0.01 |
AMZ2 |
archaelysin family metallopeptidase 2 |
27 |
0.83 |
| chr2_26367902_26368120 | 0.01 |
GAREML |
GRB2 associated, regulator of MAPK1-like |
27949 |
0.17 |
| chr5_114632718_114632869 | 0.01 |
CCDC112 |
coiled-coil domain containing 112 |
265 |
0.92 |
| chr4_2802836_2803519 | 0.01 |
SH3BP2 |
SH3-domain binding protein 2 |
2452 |
0.3 |
| chr19_49977180_49977368 | 0.01 |
FLT3LG |
fms-related tyrosine kinase 3 ligand |
211 |
0.79 |
| chr14_58893255_58894222 | 0.01 |
TIMM9 |
translocase of inner mitochondrial membrane 9 homolog (yeast) |
94 |
0.8 |
| chr16_1772321_1772575 | 0.01 |
LA16c-329F2.1 |
|
8239 |
0.08 |
| chr7_10978872_10979727 | 0.01 |
NDUFA4 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa |
584 |
0.8 |
| chr12_29301686_29302400 | 0.01 |
FAR2 |
fatty acyl CoA reductase 2 |
7 |
0.99 |
| chr5_43121819_43122643 | 0.01 |
ZNF131 |
zinc finger protein 131 |
90 |
0.97 |
| chr5_132299396_132299679 | 0.01 |
AFF4 |
AF4/FMR2 family, member 4 |
211 |
0.92 |
| chr19_54975667_54976253 | 0.01 |
LENG9 |
leukocyte receptor cluster (LRC) member 9 |
1066 |
0.31 |
| chr13_50701261_50702144 | 0.01 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
45395 |
0.14 |
| chr2_42588838_42589158 | 0.01 |
COX7A2L |
cytochrome c oxidase subunit VIIa polypeptide 2 like |
642 |
0.78 |
| chr12_132293323_132293474 | 0.01 |
ENSG00000200483 |
. |
6515 |
0.17 |
| chr4_41992555_41992786 | 0.01 |
SLC30A9 |
solute carrier family 30 (zinc transporter), member 9 |
181 |
0.96 |
| chr4_103265993_103266510 | 0.01 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
45 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |