Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
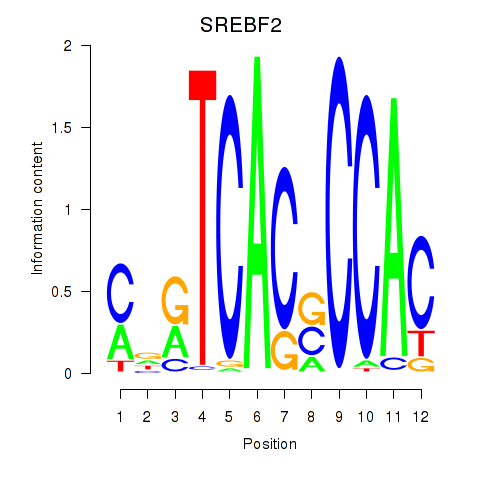
Results for SREBF2
Z-value: 0.75
Transcription factors associated with SREBF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SREBF2
|
ENSG00000198911.7 | sterol regulatory element binding transcription factor 2 |
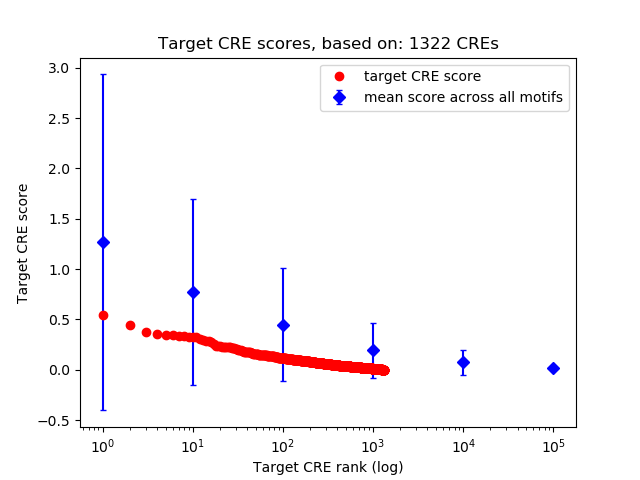
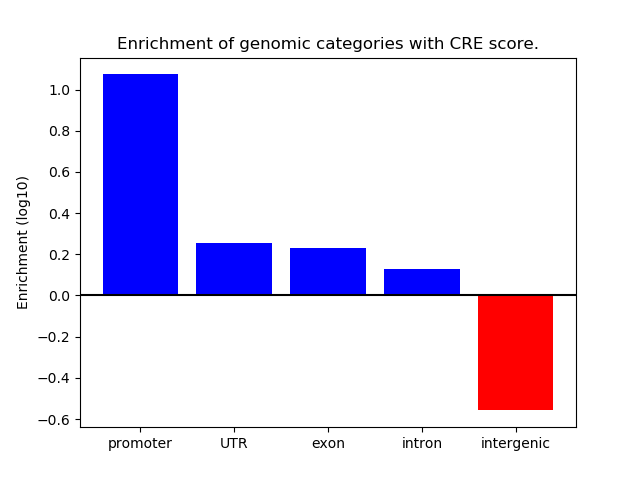
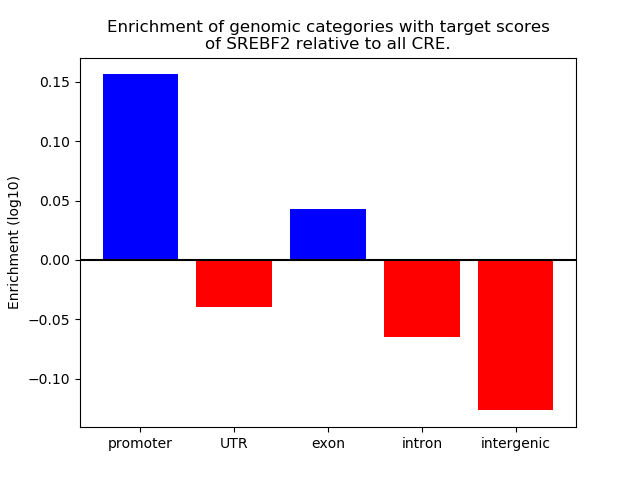
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr22_42228231_42228382 | SREBF2 | 803 | 0.480885 | 0.32 | 4.0e-01 | Click! |
| chr22_42229173_42229736 | SREBF2 | 345 | 0.756595 | -0.19 | 6.3e-01 | Click! |
| chr22_42228451_42228869 | SREBF2 | 449 | 0.708700 | 0.11 | 7.9e-01 | Click! |
Activity of the SREBF2 motif across conditions
Conditions sorted by the z-value of the SREBF2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
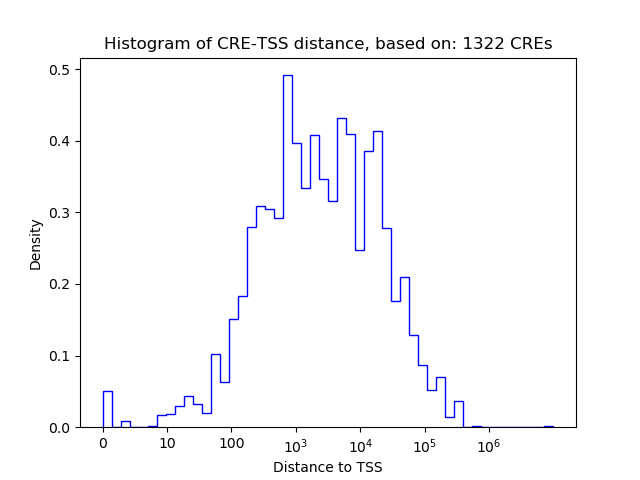
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_67172347_67172531 | 0.55 |
TBC1D10C |
TBC1 domain family, member 10C |
779 |
0.35 |
| chr17_56408210_56408765 | 0.45 |
MIR142 |
microRNA 142 |
192 |
0.87 |
| chr17_2680216_2680485 | 0.38 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
0 |
0.97 |
| chr7_1978940_1979381 | 0.36 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
1025 |
0.64 |
| chr3_111851121_111851374 | 0.35 |
GCSAM |
germinal center-associated, signaling and motility |
825 |
0.44 |
| chr20_58631308_58631666 | 0.34 |
C20orf197 |
chromosome 20 open reading frame 197 |
507 |
0.85 |
| chr1_241715191_241715459 | 0.34 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
16610 |
0.2 |
| chrX_49117374_49117525 | 0.34 |
FOXP3 |
forkhead box P3 |
1752 |
0.19 |
| chr17_79261408_79261559 | 0.33 |
SLC38A10 |
solute carrier family 38, member 10 |
5419 |
0.14 |
| chr2_113932645_113932882 | 0.33 |
AC016683.5 |
|
151 |
0.91 |
| chr4_6918168_6918454 | 0.33 |
TBC1D14 |
TBC1 domain family, member 14 |
6336 |
0.19 |
| chr7_50356004_50356399 | 0.31 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
7883 |
0.3 |
| chr17_38720511_38720770 | 0.30 |
CCR7 |
chemokine (C-C motif) receptor 7 |
1071 |
0.49 |
| chr5_171600666_171600940 | 0.29 |
STK10 |
serine/threonine kinase 10 |
14587 |
0.19 |
| chr21_47035971_47036122 | 0.28 |
PCBP3 |
poly(rC) binding protein 3 |
27562 |
0.2 |
| chr10_127688132_127688283 | 0.27 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
4221 |
0.25 |
| chr5_36725036_36725307 | 0.26 |
CTD-2353F22.1 |
|
104 |
0.98 |
| chr21_36418295_36418575 | 0.24 |
RUNX1 |
runt-related transcription factor 1 |
3027 |
0.41 |
| chr4_84034679_84034947 | 0.24 |
PLAC8 |
placenta-specific 8 |
1055 |
0.6 |
| chr3_112215334_112215665 | 0.23 |
BTLA |
B and T lymphocyte associated |
2706 |
0.33 |
| chr17_72211874_72212025 | 0.23 |
TTYH2 |
tweety family member 2 |
194 |
0.91 |
| chr9_139925276_139925479 | 0.23 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
2085 |
0.1 |
| chr22_37616154_37616305 | 0.23 |
SSTR3 |
somatostatin receptor 3 |
7867 |
0.13 |
| chr22_18260050_18260218 | 0.22 |
BID |
BH3 interacting domain death agonist |
2703 |
0.26 |
| chr2_38607378_38607679 | 0.22 |
ATL2 |
atlastin GTPase 2 |
3101 |
0.37 |
| chr19_1040639_1041023 | 0.22 |
ABCA7 |
ATP-binding cassette, sub-family A (ABC1), member 7 |
370 |
0.68 |
| chr12_6570416_6570672 | 0.22 |
VAMP1 |
vesicle-associated membrane protein 1 (synaptobrevin 1) |
9267 |
0.08 |
| chr12_111099649_111099800 | 0.22 |
TCTN1 |
tectonic family member 1 |
20870 |
0.13 |
| chr1_31228309_31228562 | 0.21 |
LAPTM5 |
lysosomal protein transmembrane 5 |
2232 |
0.26 |
| chr19_7413904_7414409 | 0.21 |
CTB-133G6.1 |
|
308 |
0.88 |
| chr20_31099035_31099434 | 0.21 |
C20orf112 |
chromosome 20 open reading frame 112 |
24 |
0.98 |
| chr4_3303508_3303659 | 0.20 |
RGS12 |
regulator of G-protein signaling 12 |
7179 |
0.25 |
| chr2_8519851_8520002 | 0.20 |
AC011747.7 |
|
295970 |
0.01 |
| chr3_101504129_101504461 | 0.19 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
51 |
0.97 |
| chr3_121378552_121379115 | 0.19 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
912 |
0.5 |
| chr17_70715496_70715723 | 0.18 |
ENSG00000222545 |
. |
4999 |
0.28 |
| chr12_4381475_4382675 | 0.18 |
CCND2 |
cyclin D2 |
863 |
0.52 |
| chr12_10096266_10096511 | 0.18 |
CLEC12A |
C-type lectin domain family 12, member A |
7527 |
0.14 |
| chr3_5019013_5019527 | 0.18 |
BHLHE40 |
basic helix-loop-helix family, member e40 |
1531 |
0.38 |
| chr10_126417275_126417426 | 0.18 |
FAM53B |
family with sequence similarity 53, member B |
14189 |
0.16 |
| chr16_88748267_88748717 | 0.18 |
SNAI3-AS1 |
SNAI3 antisense RNA 1 |
3049 |
0.1 |
| chr22_39638272_39638423 | 0.18 |
PDGFB |
platelet-derived growth factor beta polypeptide |
674 |
0.63 |
| chr20_23064979_23065243 | 0.17 |
CD93 |
CD93 molecule |
1866 |
0.32 |
| chr8_38216839_38216990 | 0.17 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
21425 |
0.11 |
| chr11_67036883_67037040 | 0.17 |
ADRBK1 |
adrenergic, beta, receptor kinase 1 |
3009 |
0.16 |
| chr15_63798871_63799191 | 0.17 |
USP3 |
ubiquitin specific peptidase 3 |
2176 |
0.37 |
| chr22_37542366_37542517 | 0.16 |
IL2RB |
interleukin 2 receptor, beta |
3589 |
0.14 |
| chr2_197047793_197047944 | 0.16 |
STK17B |
serine/threonine kinase 17b |
6641 |
0.2 |
| chr11_44326896_44327745 | 0.16 |
ALX4 |
ALX homeobox 4 |
4396 |
0.34 |
| chr12_104856651_104856876 | 0.16 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
5984 |
0.3 |
| chr2_10169783_10169983 | 0.15 |
KLF11 |
Kruppel-like factor 11 |
13093 |
0.13 |
| chr5_169724277_169724686 | 0.15 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
750 |
0.71 |
| chr15_40397944_40398095 | 0.15 |
BMF |
Bcl2 modifying factor |
268 |
0.9 |
| chr21_45341679_45341830 | 0.15 |
ENSG00000199598 |
. |
1972 |
0.29 |
| chr19_8274834_8275489 | 0.15 |
CERS4 |
ceramide synthase 4 |
643 |
0.68 |
| chr6_33610158_33610417 | 0.15 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
21126 |
0.13 |
| chr1_158901311_158901684 | 0.15 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
139 |
0.97 |
| chr8_38663717_38663929 | 0.15 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
855 |
0.58 |
| chr7_1550821_1550972 | 0.15 |
AC102953.6 |
|
1833 |
0.26 |
| chr17_29644174_29644325 | 0.15 |
CTD-2370N5.3 |
|
1409 |
0.29 |
| chr14_75085047_75085411 | 0.15 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
5923 |
0.21 |
| chr1_19253687_19254204 | 0.15 |
IFFO2 |
intermediate filament family orphan 2 |
15459 |
0.16 |
| chr14_99699001_99699256 | 0.15 |
AL109767.1 |
|
30157 |
0.18 |
| chr17_45814129_45814344 | 0.15 |
TBX21 |
T-box 21 |
3626 |
0.18 |
| chr2_43401435_43401586 | 0.14 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
52238 |
0.14 |
| chr7_105711766_105712098 | 0.14 |
SYPL1 |
synaptophysin-like 1 |
26376 |
0.22 |
| chr9_139923953_139924104 | 0.14 |
ABCA2 |
ATP-binding cassette, sub-family A (ABC1), member 2 |
1288 |
0.16 |
| chr10_126916601_126916752 | 0.14 |
CTBP2 |
C-terminal binding protein 2 |
67046 |
0.13 |
| chr19_7770859_7771101 | 0.14 |
FCER2 |
Fc fragment of IgE, low affinity II, receptor for (CD23) |
3948 |
0.11 |
| chr9_71783775_71784020 | 0.14 |
TJP2 |
tight junction protein 2 |
5184 |
0.29 |
| chr1_150532769_150532920 | 0.14 |
ADAMTSL4-AS1 |
ADAMTSL4 antisense RNA 1 |
1125 |
0.26 |
| chr6_34360722_34361201 | 0.14 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
510 |
0.78 |
| chr22_39493450_39493612 | 0.14 |
APOBEC3H |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
251 |
0.88 |
| chr14_22955216_22955367 | 0.14 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
880 |
0.4 |
| chrX_153094909_153095060 | 0.14 |
PDZD4 |
PDZ domain containing 4 |
367 |
0.75 |
| chrX_110366573_110366760 | 0.14 |
PAK3 |
p21 protein (Cdc42/Rac)-activated kinase 3 |
361 |
0.93 |
| chr1_198651611_198651863 | 0.13 |
RP11-553K8.5 |
|
15547 |
0.24 |
| chr1_12124202_12124372 | 0.13 |
TNFRSF8 |
tumor necrosis factor receptor superfamily, member 8 |
760 |
0.56 |
| chr7_41921705_41921953 | 0.13 |
AC005027.3 |
|
176896 |
0.03 |
| chr19_39825870_39826313 | 0.13 |
GMFG |
glia maturation factor, gamma |
554 |
0.58 |
| chr2_232571958_232572632 | 0.13 |
PTMA |
prothymosin, alpha |
99 |
0.91 |
| chr22_31886169_31886386 | 0.13 |
EIF4ENIF1 |
eukaryotic translation initiation factor 4E nuclear import factor 1 |
354 |
0.81 |
| chr5_909418_909569 | 0.13 |
TRIP13 |
thyroid hormone receptor interactor 13 |
15736 |
0.15 |
| chr16_56691251_56692352 | 0.13 |
MT1F |
metallothionein 1F |
54 |
0.92 |
| chr8_27183780_27184189 | 0.13 |
PTK2B |
protein tyrosine kinase 2 beta |
356 |
0.89 |
| chr4_2800321_2800472 | 0.13 |
SH3BP2 |
SH3-domain binding protein 2 |
105 |
0.97 |
| chr8_95956181_95956332 | 0.12 |
TP53INP1 |
tumor protein p53 inducible nuclear protein 1 |
3055 |
0.18 |
| chr17_47840030_47840441 | 0.12 |
FAM117A |
family with sequence similarity 117, member A |
1258 |
0.41 |
| chr6_41998934_41999085 | 0.12 |
ENSG00000206875 |
. |
12757 |
0.15 |
| chr12_123513254_123513405 | 0.12 |
PITPNM2 |
phosphatidylinositol transfer protein, membrane-associated 2 |
5872 |
0.15 |
| chr5_126094279_126094440 | 0.12 |
ENSG00000252185 |
. |
3250 |
0.29 |
| chr6_27661627_27662566 | 0.12 |
ENSG00000238648 |
. |
62908 |
0.08 |
| chr12_9911079_9911493 | 0.12 |
CD69 |
CD69 molecule |
2211 |
0.26 |
| chr19_2620958_2621288 | 0.12 |
CTC-265F19.2 |
|
9263 |
0.14 |
| chr7_100223660_100223811 | 0.12 |
TFR2 |
transferrin receptor 2 |
6992 |
0.09 |
| chr11_122567611_122567762 | 0.12 |
ENSG00000239079 |
. |
29333 |
0.19 |
| chr18_60087506_60087670 | 0.12 |
RP11-640A1.3 |
|
42225 |
0.16 |
| chr18_20651331_20651482 | 0.12 |
ENSG00000223023 |
. |
46847 |
0.11 |
| chr15_92392813_92392964 | 0.12 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
4037 |
0.29 |
| chr12_131272457_131272715 | 0.11 |
ENSG00000238822 |
. |
27312 |
0.17 |
| chr11_117856643_117857627 | 0.11 |
IL10RA |
interleukin 10 receptor, alpha |
26 |
0.98 |
| chr2_74743098_74743363 | 0.11 |
TLX2 |
T-cell leukemia homeobox 2 |
1619 |
0.14 |
| chr1_160680158_160680309 | 0.11 |
CD48 |
CD48 molecule |
1360 |
0.38 |
| chr1_1141210_1141593 | 0.11 |
TNFRSF18 |
tumor necrosis factor receptor superfamily, member 18 |
341 |
0.72 |
| chr19_14277944_14278210 | 0.11 |
LPHN1 |
latrophilin 1 |
5729 |
0.13 |
| chr9_130319902_130320178 | 0.11 |
FAM129B |
family with sequence similarity 129, member B |
11327 |
0.17 |
| chr9_123682878_123683088 | 0.11 |
TRAF1 |
TNF receptor-associated factor 1 |
6133 |
0.22 |
| chrX_78621891_78622920 | 0.11 |
ITM2A |
integral membrane protein 2A |
451 |
0.91 |
| chr5_95152547_95152698 | 0.11 |
GLRX |
glutaredoxin (thioltransferase) |
5793 |
0.16 |
| chr2_8627682_8628105 | 0.11 |
AC011747.7 |
|
188003 |
0.03 |
| chrX_39501672_39501823 | 0.11 |
ENSG00000263730 |
. |
18723 |
0.29 |
| chr21_44820304_44820455 | 0.11 |
SIK1 |
salt-inducible kinase 1 |
26629 |
0.24 |
| chr1_22463869_22464020 | 0.11 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
5515 |
0.23 |
| chr7_50350295_50350528 | 0.11 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
2093 |
0.45 |
| chr18_77159141_77159292 | 0.11 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
1106 |
0.59 |
| chr9_95895594_95896005 | 0.11 |
NINJ1 |
ninjurin 1 |
771 |
0.63 |
| chr12_122237162_122237447 | 0.11 |
RHOF |
ras homolog family member F (in filopodia) |
1279 |
0.39 |
| chr1_155231583_155232074 | 0.11 |
SCAMP3 |
secretory carrier membrane protein 3 |
222 |
0.81 |
| chr6_119634064_119634215 | 0.11 |
ENSG00000200732 |
. |
14307 |
0.25 |
| chr14_100770611_100770762 | 0.11 |
SLC25A29 |
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
2092 |
0.14 |
| chr15_75082884_75083035 | 0.10 |
ENSG00000264386 |
. |
1861 |
0.2 |
| chr18_77161034_77161555 | 0.10 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
902 |
0.67 |
| chr17_57515063_57515214 | 0.10 |
RP11-567L7.5 |
|
34474 |
0.15 |
| chr6_45345006_45345267 | 0.10 |
SUPT3H |
suppressor of Ty 3 homolog (S. cerevisiae) |
534 |
0.83 |
| chr17_76771045_76771282 | 0.10 |
CYTH1 |
cytohesin 1 |
7191 |
0.19 |
| chr12_48398455_48398676 | 0.10 |
COL2A1 |
collagen, type II, alpha 1 |
296 |
0.86 |
| chr16_68390170_68390671 | 0.10 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
14488 |
0.11 |
| chr7_1051859_1052010 | 0.10 |
ENSG00000199023 |
. |
10728 |
0.1 |
| chr3_128523693_128523920 | 0.10 |
RAB7A |
RAB7A, member RAS oncogene family |
9604 |
0.18 |
| chr9_132222026_132222209 | 0.10 |
ENSG00000264298 |
. |
18718 |
0.2 |
| chr6_167507024_167507239 | 0.10 |
CCR6 |
chemokine (C-C motif) receptor 6 |
18164 |
0.18 |
| chr3_71478867_71479018 | 0.10 |
ENSG00000221264 |
. |
112298 |
0.06 |
| chr17_80063977_80064793 | 0.10 |
CCDC57 |
coiled-coil domain containing 57 |
4659 |
0.1 |
| chr9_134113693_134114226 | 0.10 |
NUP214 |
nucleoporin 214kDa |
10278 |
0.17 |
| chr17_48217501_48218008 | 0.10 |
AC002401.1 |
|
4803 |
0.11 |
| chr22_25960035_25960186 | 0.10 |
CTA-407F11.8 |
|
7 |
0.93 |
| chr2_143885996_143886303 | 0.10 |
ARHGAP15 |
Rho GTPase activating protein 15 |
734 |
0.76 |
| chr1_47048198_47048369 | 0.10 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
2949 |
0.2 |
| chr12_12867727_12867916 | 0.10 |
CDKN1B |
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
2237 |
0.23 |
| chr3_183852600_183853095 | 0.10 |
RP11-778D9.12 |
|
122 |
0.61 |
| chr22_44578201_44578448 | 0.10 |
PARVG |
parvin, gamma |
583 |
0.83 |
| chr17_80229441_80229710 | 0.10 |
CSNK1D |
casein kinase 1, delta |
1729 |
0.23 |
| chr3_14241386_14241537 | 0.10 |
XPC |
xeroderma pigmentosum, complementation group C |
21178 |
0.17 |
| chr13_37574524_37574865 | 0.10 |
EXOSC8 |
exosome component 8 |
16 |
0.96 |
| chr3_128734717_128734917 | 0.10 |
EFCC1 |
EF-hand and coiled-coil domain containing 1 |
14345 |
0.13 |
| chr5_156700321_156700472 | 0.10 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
4034 |
0.17 |
| chr8_134071502_134071774 | 0.10 |
SLA |
Src-like-adaptor |
965 |
0.65 |
| chr22_38036611_38036887 | 0.10 |
SH3BP1 |
SH3-domain binding protein 1 |
722 |
0.49 |
| chr2_233925739_233925978 | 0.10 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
669 |
0.7 |
| chr9_5832277_5833455 | 0.10 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
251 |
0.93 |
| chr10_25176560_25176711 | 0.09 |
ENSG00000240294 |
. |
21106 |
0.23 |
| chr10_106083582_106083733 | 0.09 |
ITPRIP |
inositol 1,4,5-trisphosphate receptor interacting protein |
5025 |
0.15 |
| chr1_154379631_154380335 | 0.09 |
RP11-350G8.5 |
|
943 |
0.42 |
| chr2_204572488_204572658 | 0.09 |
CD28 |
CD28 molecule |
1157 |
0.58 |
| chr20_57266602_57266883 | 0.09 |
NPEPL1 |
aminopeptidase-like 1 |
927 |
0.53 |
| chr9_130546021_130546292 | 0.09 |
CDK9 |
cyclin-dependent kinase 9 |
1802 |
0.16 |
| chr1_44873108_44873259 | 0.09 |
RNF220 |
ring finger protein 220 |
2223 |
0.29 |
| chr7_154997328_154997479 | 0.09 |
AC099552.4 |
Uncharacterized protein |
7257 |
0.22 |
| chr14_23341701_23341904 | 0.09 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
289 |
0.77 |
| chr17_9549466_9549768 | 0.09 |
USP43 |
ubiquitin specific peptidase 43 |
763 |
0.39 |
| chr14_35303482_35303633 | 0.09 |
ENSG00000251726 |
. |
11009 |
0.17 |
| chr16_70286070_70286221 | 0.09 |
EXOSC6 |
exosome component 6 |
312 |
0.77 |
| chr13_91827106_91827257 | 0.09 |
ENSG00000215417 |
. |
175678 |
0.04 |
| chr12_68775706_68775857 | 0.09 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
49620 |
0.17 |
| chr11_112096740_112097015 | 0.09 |
PTS |
6-pyruvoyltetrahydropterin synthase |
211 |
0.89 |
| chr1_12244133_12244323 | 0.09 |
ENSG00000263676 |
. |
7542 |
0.16 |
| chrX_23872431_23872582 | 0.09 |
APOO |
apolipoprotein O |
26512 |
0.15 |
| chr3_112323750_112323901 | 0.09 |
CCDC80 |
coiled-coil domain containing 80 |
5302 |
0.24 |
| chr7_102044904_102045055 | 0.09 |
ENSG00000221510 |
. |
1323 |
0.27 |
| chr2_85806418_85806569 | 0.09 |
VAMP8 |
vesicle-associated membrane protein 8 |
1777 |
0.17 |
| chr13_78272907_78273541 | 0.09 |
SLAIN1 |
SLAIN motif family, member 1 |
207 |
0.89 |
| chr8_131020590_131020791 | 0.09 |
ENSG00000264653 |
. |
9 |
0.97 |
| chr9_140171799_140172072 | 0.09 |
TOR4A |
torsin family 4, member A |
266 |
0.79 |
| chr9_92786212_92786363 | 0.09 |
ENSG00000263967 |
. |
470 |
0.9 |
| chr2_241390296_241390447 | 0.09 |
GPC1 |
glypican 1 |
1857 |
0.28 |
| chr1_226920663_226921046 | 0.09 |
ITPKB |
inositol-trisphosphate 3-kinase B |
4305 |
0.27 |
| chr4_75263884_75264078 | 0.09 |
EREG |
epiregulin |
33121 |
0.14 |
| chr10_90754998_90755312 | 0.09 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
4008 |
0.17 |
| chr16_31411645_31411796 | 0.09 |
RP11-120K18.2 |
|
2166 |
0.18 |
| chr6_15245727_15246152 | 0.09 |
JARID2 |
jumonji, AT rich interactive domain 2 |
588 |
0.74 |
| chr3_13462664_13462815 | 0.09 |
NUP210 |
nucleoporin 210kDa |
930 |
0.65 |
| chr22_22293537_22293770 | 0.08 |
PPM1F |
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
697 |
0.45 |
| chr9_139929070_139929505 | 0.08 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
1825 |
0.11 |
| chr9_79265748_79266757 | 0.08 |
PRUNE2 |
prune homolog 2 (Drosophila) |
1213 |
0.53 |
| chr3_71751711_71751862 | 0.08 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
22740 |
0.2 |
| chr22_20225722_20225873 | 0.08 |
RTN4R |
reticulon 4 receptor |
5410 |
0.14 |
| chr2_233179871_233180040 | 0.08 |
DIS3L2 |
DIS3 mitotic control homolog (S. cerevisiae)-like 2 |
18698 |
0.17 |
| chr18_72166510_72167447 | 0.08 |
CNDP2 |
CNDP dipeptidase 2 (metallopeptidase M20 family) |
118 |
0.97 |
| chr1_111174065_111174216 | 0.08 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
44 |
0.98 |
| chr17_38769482_38769633 | 0.08 |
SMARCE1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
18985 |
0.13 |
| chr12_124950039_124950191 | 0.08 |
NCOR2 |
nuclear receptor corepressor 2 |
18200 |
0.28 |
| chr11_67857787_67857938 | 0.08 |
CHKA |
choline kinase alpha |
30433 |
0.1 |
| chr12_122239467_122240363 | 0.08 |
RHOF |
ras homolog family member F (in filopodia) |
919 |
0.49 |
| chr17_75450826_75450977 | 0.08 |
SEPT9 |
septin 9 |
739 |
0.6 |
| chr10_53458598_53459393 | 0.08 |
CSTF2T |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant |
360 |
0.92 |
| chr2_144941449_144941842 | 0.08 |
GTDC1 |
glycosyltransferase-like domain containing 1 |
51659 |
0.18 |
| chr1_19805004_19805155 | 0.08 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
5500 |
0.18 |
| chr1_200989934_200990087 | 0.08 |
KIF21B |
kinesin family member 21B |
2526 |
0.27 |
| chr17_36860259_36860862 | 0.08 |
MLLT6 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
1235 |
0.2 |
| chr19_41857713_41858218 | 0.08 |
TMEM91 |
transmembrane protein 91 |
1149 |
0.27 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.2 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.0 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |