Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
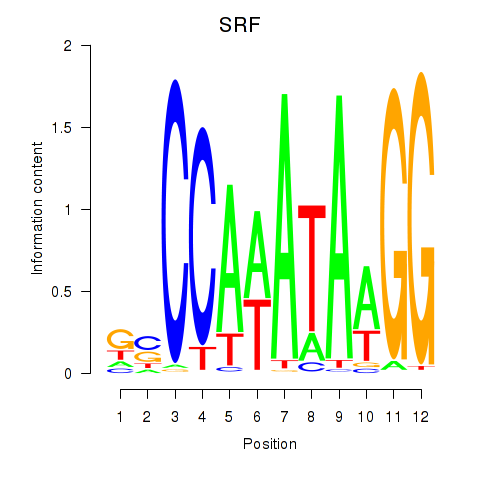
Results for SRF
Z-value: 0.96
Transcription factors associated with SRF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRF
|
ENSG00000112658.6 | serum response factor |
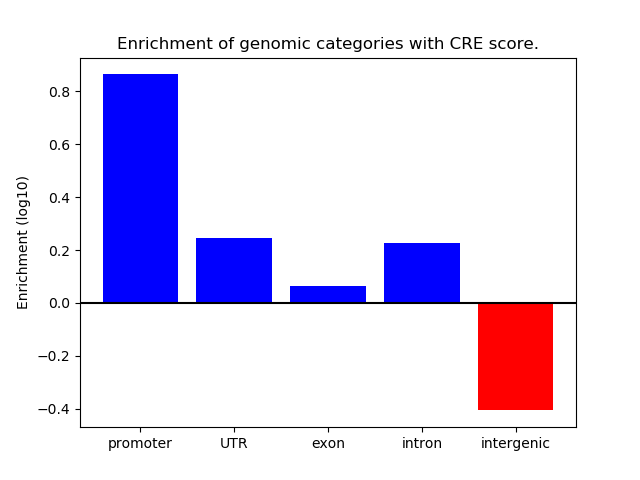
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_43143818_43143969 | SRF | 3765 | 0.148525 | 0.64 | 6.2e-02 | Click! |
| chr6_43141770_43142073 | SRF | 1793 | 0.242388 | 0.50 | 1.7e-01 | Click! |
| chr6_43140857_43141008 | SRF | 804 | 0.508783 | 0.41 | 2.8e-01 | Click! |
| chr6_43142093_43142405 | SRF | 2121 | 0.210632 | 0.33 | 3.9e-01 | Click! |
| chr6_43142593_43142764 | SRF | 2550 | 0.184052 | 0.25 | 5.1e-01 | Click! |
Activity of the SRF motif across conditions
Conditions sorted by the z-value of the SRF motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
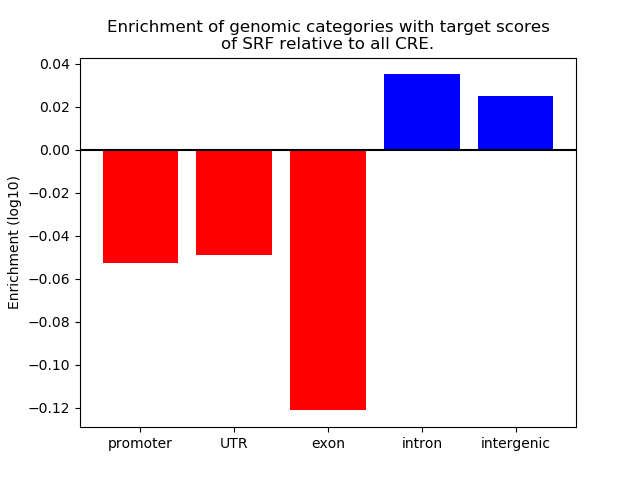
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_103048383_103048621 | 0.58 |
PLP1 |
proteolipid protein 1 |
16724 |
0.13 |
| chr7_28727324_28727475 | 0.54 |
CREB5 |
cAMP responsive element binding protein 5 |
1801 |
0.52 |
| chr5_92918344_92919004 | 0.52 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
369 |
0.84 |
| chr2_118737624_118738057 | 0.45 |
AC009303.1 |
|
15133 |
0.14 |
| chr3_25470006_25470271 | 0.45 |
RARB |
retinoic acid receptor, beta |
336 |
0.93 |
| chr1_12588381_12588669 | 0.44 |
ENSG00000239149 |
. |
21225 |
0.17 |
| chr5_2750019_2750705 | 0.43 |
IRX2 |
iroquois homeobox 2 |
1407 |
0.43 |
| chr11_6340282_6340518 | 0.43 |
PRKCDBP |
protein kinase C, delta binding protein |
1362 |
0.38 |
| chr15_26020644_26020846 | 0.42 |
ENSG00000222123 |
. |
37090 |
0.15 |
| chr3_64674409_64674678 | 0.41 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
867 |
0.65 |
| chr21_36255325_36255652 | 0.41 |
RUNX1 |
runt-related transcription factor 1 |
3992 |
0.36 |
| chr6_108434801_108435121 | 0.40 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
39020 |
0.14 |
| chr3_157250255_157250502 | 0.39 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
1017 |
0.61 |
| chr15_39871853_39872515 | 0.38 |
THBS1 |
thrombospondin 1 |
1096 |
0.52 |
| chr3_25193145_25193356 | 0.38 |
RARB |
retinoic acid receptor, beta |
22573 |
0.26 |
| chr16_15952010_15952161 | 0.37 |
MYH11 |
myosin, heavy chain 11, smooth muscle |
1195 |
0.43 |
| chr6_2376939_2377282 | 0.37 |
ENSG00000266252 |
. |
32759 |
0.23 |
| chr7_139542200_139542351 | 0.36 |
TBXAS1 |
thromboxane A synthase 1 (platelet) |
13165 |
0.26 |
| chr8_86350363_86351282 | 0.35 |
CA3 |
carbonic anhydrase III, muscle specific |
234 |
0.93 |
| chr4_170188672_170188986 | 0.35 |
SH3RF1 |
SH3 domain containing ring finger 1 |
2279 |
0.41 |
| chr15_51401233_51401466 | 0.33 |
TNFAIP8L3 |
tumor necrosis factor, alpha-induced protein 8-like 3 |
3876 |
0.27 |
| chr1_154163082_154163233 | 0.32 |
TPM3 |
tropomyosin 3 |
1377 |
0.24 |
| chr7_34027132_34027307 | 0.32 |
BMPER |
BMP binding endothelial regulator |
82074 |
0.11 |
| chr12_121086851_121087153 | 0.32 |
CABP1 |
calcium binding protein 1 |
886 |
0.46 |
| chr1_156098945_156099118 | 0.32 |
LMNA |
lamin A/C |
2437 |
0.17 |
| chr12_110547123_110547283 | 0.31 |
IFT81 |
intraflagellar transport 81 homolog (Chlamydomonas) |
14937 |
0.2 |
| chr7_68895099_68895250 | 0.31 |
ENSG00000252423 |
. |
28885 |
0.22 |
| chr3_99679660_99679890 | 0.31 |
ENSG00000264897 |
. |
3467 |
0.34 |
| chr2_12927970_12928158 | 0.31 |
ENSG00000264370 |
. |
50571 |
0.17 |
| chr8_89338891_89339093 | 0.29 |
RP11-586K2.1 |
|
73 |
0.93 |
| chr1_223889194_223889423 | 0.29 |
CAPN2 |
calpain 2, (m/II) large subunit |
13 |
0.98 |
| chr15_35598885_35599113 | 0.29 |
ENSG00000265102 |
. |
65566 |
0.14 |
| chr8_26123485_26124097 | 0.29 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
25216 |
0.23 |
| chr15_68871678_68872325 | 0.29 |
CORO2B |
coronin, actin binding protein, 2B |
428 |
0.9 |
| chr10_29923748_29924013 | 0.28 |
SVIL |
supervillin |
21 |
0.98 |
| chr14_56792096_56792462 | 0.28 |
TMEM260 |
transmembrane protein 260 |
162793 |
0.04 |
| chr4_89977525_89978081 | 0.28 |
FAM13A |
family with sequence similarity 13, member A |
508 |
0.85 |
| chr1_180126514_180126782 | 0.28 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
2616 |
0.34 |
| chr1_157964321_157964712 | 0.28 |
KIRREL |
kin of IRRE like (Drosophila) |
1081 |
0.56 |
| chr8_119961799_119961950 | 0.27 |
TNFRSF11B |
tumor necrosis factor receptor superfamily, member 11b |
2565 |
0.34 |
| chr5_139145492_139145717 | 0.27 |
PSD2 |
pleckstrin and Sec7 domain containing 2 |
29802 |
0.16 |
| chrX_100334578_100334939 | 0.27 |
TMEM35 |
transmembrane protein 35 |
1049 |
0.47 |
| chr6_78172796_78173983 | 0.27 |
HTR1B |
5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
101 |
0.99 |
| chr5_142875468_142875722 | 0.26 |
ENSG00000253023 |
. |
46557 |
0.16 |
| chr12_54414044_54414195 | 0.26 |
HOXC4 |
homeobox C4 |
3404 |
0.08 |
| chr10_17256481_17257128 | 0.26 |
VIM-AS1 |
VIM antisense RNA 1 |
12093 |
0.15 |
| chr5_120534956_120535178 | 0.26 |
ENSG00000222609 |
. |
488532 |
0.01 |
| chr12_115113843_115113994 | 0.26 |
TBX3 |
T-box 3 |
7477 |
0.23 |
| chr2_208578578_208578729 | 0.25 |
CCNYL1 |
cyclin Y-like 1 |
2178 |
0.25 |
| chr12_112534683_112534834 | 0.25 |
NAA25 |
N(alpha)-acetyltransferase 25, NatB auxiliary subunit |
11789 |
0.13 |
| chrX_80514661_80514812 | 0.25 |
ENSG00000264374 |
. |
18417 |
0.23 |
| chr12_109123891_109124402 | 0.25 |
CORO1C |
coronin, actin binding protein, 1C |
181 |
0.94 |
| chr2_161236822_161236973 | 0.25 |
ENSG00000252465 |
. |
16582 |
0.2 |
| chr14_34161873_34162024 | 0.25 |
NPAS3 |
neuronal PAS domain protein 3 |
42499 |
0.22 |
| chrX_153601896_153602918 | 0.25 |
FLNA |
filamin A, alpha |
587 |
0.54 |
| chr15_63179528_63179679 | 0.24 |
RP11-1069G10.1 |
|
3370 |
0.3 |
| chr10_33621519_33621932 | 0.24 |
NRP1 |
neuropilin 1 |
1585 |
0.49 |
| chr5_76786577_76786860 | 0.24 |
WDR41 |
WD repeat domain 41 |
392 |
0.91 |
| chr18_57488899_57489050 | 0.24 |
ENSG00000221471 |
. |
72065 |
0.1 |
| chr21_28215844_28216573 | 0.24 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
112 |
0.98 |
| chr3_160191571_160191722 | 0.24 |
ENSG00000238427 |
. |
1442 |
0.35 |
| chr1_95332163_95332314 | 0.24 |
SLC44A3 |
solute carrier family 44, member 3 |
649 |
0.72 |
| chr16_84328151_84329018 | 0.24 |
WFDC1 |
WAP four-disulfide core domain 1 |
132 |
0.94 |
| chr8_97398942_97399239 | 0.24 |
ENSG00000202095 |
. |
44298 |
0.15 |
| chr9_97818122_97818533 | 0.24 |
ENSG00000207563 |
. |
29163 |
0.12 |
| chr6_17932450_17932601 | 0.23 |
KIF13A |
kinesin family member 13A |
55169 |
0.15 |
| chr15_70995926_70996077 | 0.23 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
387 |
0.91 |
| chr4_56264706_56264857 | 0.23 |
TMEM165 |
transmembrane protein 165 |
2651 |
0.21 |
| chr8_62625183_62625719 | 0.23 |
ASPH |
aspartate beta-hydroxylase |
1633 |
0.37 |
| chr12_15930396_15930547 | 0.23 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
3190 |
0.35 |
| chr9_122130687_122130838 | 0.23 |
BRINP1 |
bone morphogenetic protein/retinoic acid inducible neural-specific 1 |
941 |
0.74 |
| chr1_114761855_114762022 | 0.23 |
SYT6 |
synaptotagmin VI |
65397 |
0.13 |
| chr15_40632274_40632449 | 0.22 |
C15orf52 |
chromosome 15 open reading frame 52 |
118 |
0.91 |
| chr5_89351556_89351717 | 0.22 |
ENSG00000264342 |
. |
39099 |
0.23 |
| chr2_66664825_66665240 | 0.22 |
MEIS1 |
Meis homeobox 1 |
93 |
0.96 |
| chr2_241520309_241520460 | 0.22 |
RNPEPL1 |
arginyl aminopeptidase (aminopeptidase B)-like 1 |
5465 |
0.12 |
| chr3_93743277_93743462 | 0.22 |
STX19 |
syntaxin 19 |
4085 |
0.18 |
| chr5_173390424_173390761 | 0.22 |
C5orf47 |
chromosome 5 open reading frame 47 |
25570 |
0.2 |
| chr11_118435857_118436109 | 0.22 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
378 |
0.76 |
| chr3_149902419_149902570 | 0.22 |
RP11-167H9.4 |
|
86675 |
0.09 |
| chr12_104990144_104990321 | 0.21 |
ENSG00000264295 |
. |
4821 |
0.26 |
| chr8_9953745_9953924 | 0.21 |
MSRA |
methionine sulfoxide reductase A |
597 |
0.76 |
| chr4_113739998_113740296 | 0.21 |
ANK2 |
ankyrin 2, neuronal |
882 |
0.65 |
| chr3_185531595_185531973 | 0.21 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
7065 |
0.24 |
| chr1_216705335_216705486 | 0.21 |
USH2A |
Usher syndrome 2A (autosomal recessive, mild) |
108672 |
0.08 |
| chr2_26258800_26259062 | 0.21 |
RAB10 |
RAB10, member RAS oncogene family |
1952 |
0.29 |
| chr12_54426653_54426823 | 0.21 |
HOXC5 |
homeobox C5 |
101 |
0.88 |
| chr6_43141770_43142073 | 0.21 |
SRF |
serum response factor (c-fos serum response element-binding transcription factor) |
1793 |
0.24 |
| chr18_66106611_66106762 | 0.20 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
275608 |
0.02 |
| chr3_48646099_48647042 | 0.20 |
UQCRC1 |
ubiquinol-cytochrome c reductase core protein I |
900 |
0.38 |
| chr6_17283021_17283300 | 0.20 |
RBM24 |
RNA binding motif protein 24 |
228 |
0.96 |
| chr20_10651490_10651885 | 0.20 |
RP11-103J8.1 |
|
1889 |
0.38 |
| chr7_55546043_55546306 | 0.20 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
37596 |
0.19 |
| chr16_54465247_54465407 | 0.20 |
ENSG00000264079 |
. |
117981 |
0.06 |
| chr1_178837029_178837180 | 0.20 |
ANGPTL1 |
angiopoietin-like 1 |
1309 |
0.49 |
| chr3_159451123_159451337 | 0.20 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
30272 |
0.17 |
| chr8_48392520_48392671 | 0.20 |
SPIDR |
scaffolding protein involved in DNA repair |
39632 |
0.18 |
| chr6_105936815_105937042 | 0.20 |
PREP |
prolyl endopeptidase |
85969 |
0.1 |
| chr15_35087756_35087962 | 0.20 |
ACTC1 |
actin, alpha, cardiac muscle 1 |
481 |
0.83 |
| chr8_67015907_67016058 | 0.19 |
TRIM55 |
tripartite motif containing 55 |
23149 |
0.22 |
| chr21_35925267_35925598 | 0.19 |
RCAN1 |
regulator of calcineurin 1 |
7045 |
0.23 |
| chr17_76880061_76880570 | 0.19 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
10083 |
0.14 |
| chr17_36621594_36621757 | 0.19 |
ARHGAP23 |
Rho GTPase activating protein 23 |
6505 |
0.17 |
| chr19_13135028_13135217 | 0.19 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
272 |
0.84 |
| chr14_23538007_23538158 | 0.19 |
ACIN1 |
apoptotic chromatin condensation inducer 1 |
2665 |
0.13 |
| chr5_14171520_14171879 | 0.19 |
TRIO |
trio Rho guanine nucleotide exchange factor |
12208 |
0.31 |
| chr4_81118792_81119547 | 0.19 |
PRDM8 |
PR domain containing 8 |
511 |
0.82 |
| chr6_5208630_5208872 | 0.19 |
ENSG00000264541 |
. |
16279 |
0.2 |
| chr2_64539316_64539525 | 0.18 |
ENSG00000264297 |
. |
28473 |
0.21 |
| chr5_173207490_173207641 | 0.18 |
ENSG00000263401 |
. |
50484 |
0.16 |
| chr21_36254420_36254945 | 0.18 |
RUNX1 |
runt-related transcription factor 1 |
4798 |
0.34 |
| chr2_69405192_69405451 | 0.18 |
ENSG00000251850 |
. |
3814 |
0.24 |
| chr18_56665001_56665175 | 0.18 |
ENSG00000251870 |
. |
71560 |
0.1 |
| chr6_33180748_33181418 | 0.18 |
RING1 |
ring finger protein 1 |
4811 |
0.08 |
| chr11_117069900_117070051 | 0.18 |
TAGLN |
transgelin |
62 |
0.96 |
| chr9_128235766_128235975 | 0.18 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
10910 |
0.23 |
| chr5_127447715_127447866 | 0.18 |
SLC12A2 |
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
28250 |
0.26 |
| chr11_64389104_64389255 | 0.18 |
NRXN2 |
neurexin 2 |
20979 |
0.13 |
| chr4_185744284_185744435 | 0.18 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
2829 |
0.25 |
| chr11_110967368_110967519 | 0.18 |
ENSG00000200168 |
. |
56511 |
0.15 |
| chr1_98569393_98569544 | 0.18 |
ENSG00000225206 |
. |
57741 |
0.17 |
| chr9_14320711_14320862 | 0.18 |
NFIB |
nuclear factor I/B |
1551 |
0.46 |
| chr18_65661971_65662168 | 0.18 |
DSEL |
dermatan sulfate epimerase-like |
477852 |
0.01 |
| chrX_152761685_152761943 | 0.18 |
HAUS7 |
HAUS augmin-like complex, subunit 7 |
836 |
0.41 |
| chr4_55100810_55101099 | 0.17 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
4465 |
0.31 |
| chr10_97501950_97502206 | 0.17 |
ENTPD1 |
ectonucleoside triphosphate diphosphohydrolase 1 |
13331 |
0.21 |
| chr6_152701268_152701555 | 0.17 |
SYNE1-AS1 |
SYNE1 antisense RNA 1 |
270 |
0.91 |
| chr4_24796744_24797030 | 0.17 |
SOD3 |
superoxide dismutase 3, extracellular |
198 |
0.97 |
| chr20_34025363_34025770 | 0.17 |
GDF5 |
growth differentiation factor 5 |
457 |
0.71 |
| chr8_99007536_99007844 | 0.17 |
MATN2 |
matrilin 2 |
21082 |
0.15 |
| chr15_96869088_96869239 | 0.17 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
4 |
0.96 |
| chr17_58821066_58821723 | 0.17 |
ENSG00000252534 |
. |
5013 |
0.22 |
| chr4_23892235_23892469 | 0.17 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
652 |
0.82 |
| chr8_29107320_29107471 | 0.17 |
KIF13B |
kinesin family member 13B |
13195 |
0.17 |
| chr6_2468845_2468996 | 0.17 |
ENSG00000266252 |
. |
59051 |
0.15 |
| chr15_68573463_68573639 | 0.17 |
FEM1B |
fem-1 homolog b (C. elegans) |
1254 |
0.47 |
| chr16_87481635_87481786 | 0.17 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
14970 |
0.16 |
| chr14_75688794_75688945 | 0.17 |
ENSG00000252013 |
. |
19997 |
0.14 |
| chr6_3749707_3750198 | 0.17 |
RP11-420L9.5 |
|
1393 |
0.41 |
| chr18_395366_395517 | 0.17 |
RP11-720L2.2 |
|
28975 |
0.2 |
| chr2_109257559_109258085 | 0.17 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
13446 |
0.22 |
| chr17_56031961_56032485 | 0.17 |
CUEDC1 |
CUE domain containing 1 |
395 |
0.83 |
| chr5_73109422_73109633 | 0.17 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
184 |
0.95 |
| chr15_63305654_63305860 | 0.16 |
TPM1 |
tropomyosin 1 (alpha) |
29074 |
0.16 |
| chr5_150284813_150285011 | 0.16 |
ZNF300 |
zinc finger protein 300 |
367 |
0.87 |
| chrX_45613808_45613959 | 0.16 |
ENSG00000207725 |
. |
7353 |
0.25 |
| chr4_95332017_95332168 | 0.16 |
PDLIM5 |
PDZ and LIM domain 5 |
40945 |
0.2 |
| chr11_34675128_34675279 | 0.16 |
EHF |
ets homologous factor |
11029 |
0.28 |
| chr1_16212355_16212620 | 0.16 |
SPEN |
spen family transcriptional repressor |
9416 |
0.15 |
| chr6_166532528_166532679 | 0.16 |
ENSG00000231297 |
. |
19262 |
0.22 |
| chr22_27105780_27105987 | 0.16 |
CRYBA4 |
crystallin, beta A4 |
87955 |
0.08 |
| chr16_3309520_3309673 | 0.16 |
MEFV |
Mediterranean fever |
2969 |
0.14 |
| chr3_72287415_72287566 | 0.16 |
ENSG00000212070 |
. |
24089 |
0.27 |
| chr2_234151079_234151230 | 0.16 |
ATG16L1 |
autophagy related 16-like 1 (S. cerevisiae) |
9063 |
0.15 |
| chr22_39260401_39260552 | 0.16 |
CBX6 |
chromobox homolog 6 |
7745 |
0.14 |
| chr6_16849709_16849927 | 0.16 |
RP1-151F17.1 |
|
87675 |
0.09 |
| chr18_37379955_37380106 | 0.16 |
ENSG00000212354 |
. |
121444 |
0.06 |
| chr2_33391334_33391485 | 0.16 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
31685 |
0.23 |
| chrX_34673939_34674563 | 0.16 |
TMEM47 |
transmembrane protein 47 |
1154 |
0.68 |
| chr1_68215659_68215876 | 0.16 |
ENSG00000238778 |
. |
22569 |
0.2 |
| chr1_29066466_29066617 | 0.16 |
YTHDF2 |
YTH domain family, member 2 |
3064 |
0.23 |
| chr21_34677268_34677419 | 0.16 |
IFNAR1 |
interferon (alpha, beta and omega) receptor 1 |
19391 |
0.14 |
| chr2_99767240_99767391 | 0.16 |
TSGA10 |
testis specific, 10 |
4065 |
0.14 |
| chr4_186425617_186425867 | 0.16 |
PDLIM3 |
PDZ and LIM domain 3 |
30914 |
0.12 |
| chr2_20001539_20001690 | 0.16 |
TTC32 |
tetratricopeptide repeat domain 32 |
99788 |
0.08 |
| chr2_102321542_102321693 | 0.16 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
6625 |
0.32 |
| chr8_91996797_91997429 | 0.16 |
C8orf88 |
chromosome 8 open reading frame 88 |
372 |
0.85 |
| chr14_55239224_55239463 | 0.15 |
SAMD4A |
sterile alpha motif domain containing 4A |
17792 |
0.24 |
| chr8_57595049_57595246 | 0.15 |
RP11-17A4.2 |
|
193490 |
0.03 |
| chr16_1664599_1665551 | 0.15 |
CRAMP1L |
Crm, cramped-like (Drosophila) |
434 |
0.69 |
| chr20_58516772_58516923 | 0.15 |
FAM217B |
family with sequence similarity 217, member B |
332 |
0.82 |
| chr5_115313493_115313761 | 0.15 |
AQPEP |
Aminopeptidase Q |
15436 |
0.17 |
| chr2_199237110_199237329 | 0.15 |
ENSG00000252511 |
. |
194234 |
0.03 |
| chr21_15921425_15921576 | 0.15 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
2836 |
0.33 |
| chr6_138170630_138170781 | 0.15 |
RP11-356I2.4 |
|
8480 |
0.22 |
| chr3_150089259_150089633 | 0.15 |
TSC22D2 |
TSC22 domain family, member 2 |
36676 |
0.21 |
| chr6_164530063_164530741 | 0.15 |
ENSG00000266128 |
. |
268809 |
0.02 |
| chr5_146933859_146934303 | 0.15 |
DPYSL3 |
dihydropyrimidinase-like 3 |
44462 |
0.19 |
| chr1_23066485_23066803 | 0.15 |
ENSG00000216157 |
. |
8760 |
0.17 |
| chr11_63438034_63439177 | 0.15 |
ATL3 |
atlastin GTPase 3 |
479 |
0.76 |
| chr2_160418896_160419047 | 0.15 |
ENSG00000207117 |
. |
26114 |
0.19 |
| chr1_63590436_63590751 | 0.15 |
ENSG00000252259 |
. |
61528 |
0.14 |
| chr4_79473579_79473973 | 0.15 |
ANXA3 |
annexin A3 |
655 |
0.79 |
| chr2_175203738_175203889 | 0.15 |
AC018470.1 |
Uncharacterized protein FLJ46347 |
1662 |
0.32 |
| chr10_104817675_104817826 | 0.15 |
NT5C2 |
5'-nucleotidase, cytosolic II |
36375 |
0.18 |
| chr8_38264174_38264701 | 0.15 |
RP11-350N15.6 |
|
1823 |
0.22 |
| chrX_47142357_47142518 | 0.15 |
ENSG00000266158 |
. |
1975 |
0.25 |
| chr8_122419386_122419537 | 0.14 |
ENSG00000263525 |
. |
135774 |
0.05 |
| chr7_5710455_5710606 | 0.14 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
9562 |
0.21 |
| chr5_53358972_53359145 | 0.14 |
ENSG00000265421 |
. |
12355 |
0.22 |
| chr11_59021844_59021995 | 0.14 |
ENSG00000263944 |
. |
36887 |
0.12 |
| chr5_157911190_157911341 | 0.14 |
CTD-2363C16.1 |
|
498749 |
0.0 |
| chr14_21564786_21565064 | 0.14 |
ZNF219 |
zinc finger protein 219 |
1874 |
0.17 |
| chr2_85504683_85504834 | 0.14 |
ENSG00000221579 |
. |
21399 |
0.12 |
| chr12_112560647_112560960 | 0.14 |
TRAFD1 |
TRAF-type zinc finger domain containing 1 |
2502 |
0.22 |
| chrX_10851859_10852068 | 0.14 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
190 |
0.97 |
| chr9_99249166_99249317 | 0.14 |
HABP4 |
hyaluronan binding protein 4 |
36754 |
0.17 |
| chr20_61817533_61817684 | 0.14 |
ENSG00000207598 |
. |
7756 |
0.14 |
| chr1_225635412_225635563 | 0.14 |
RP11-496N12.6 |
|
17498 |
0.17 |
| chr19_11650748_11650916 | 0.14 |
CNN1 |
calponin 1, basic, smooth muscle |
123 |
0.92 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.2 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.2 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.1 | 0.2 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.1 | 0.2 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.2 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.5 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0036296 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.2 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.2 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.0 | GO:0032616 | interleukin-13 production(GO:0032616) |
| 0.0 | 0.2 | GO:0019430 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.0 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.0 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0009886 | post-embryonic morphogenesis(GO:0009886) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0031114 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0060751 | branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0033085 | negative regulation of T cell differentiation in thymus(GO:0033085) negative regulation of thymocyte aggregation(GO:2000399) |
| 0.0 | 0.0 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.5 | GO:0034339 | obsolete regulation of transcription from RNA polymerase II promoter by nuclear hormone receptor(GO:0034339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle(GO:0030990) intraciliary transport particle B(GO:0030992) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.7 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.1 | 0.5 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.0 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0017022 | myosin binding(GO:0017022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |