Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
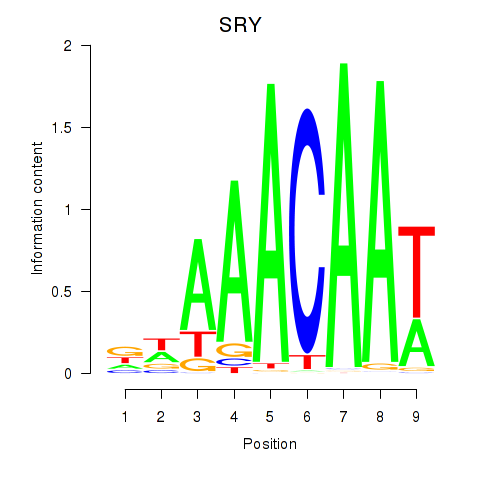
Results for SRY
Z-value: 0.25
Transcription factors associated with SRY
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SRY
|
ENSG00000184895.6 | sex determining region Y |
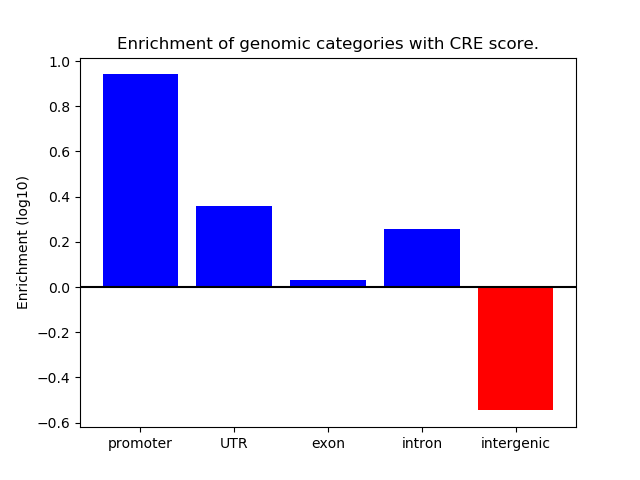
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrY_2655519_2655670 | SRY | 50 | 0.977279 | -0.81 | 7.5e-03 | Click! |
| chrY_2654987_2655138 | SRY | 582 | 0.749452 | -0.77 | 1.6e-02 | Click! |
| chrY_2655183_2655334 | SRY | 386 | 0.861759 | -0.51 | 1.6e-01 | Click! |
Activity of the SRY motif across conditions
Conditions sorted by the z-value of the SRY motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
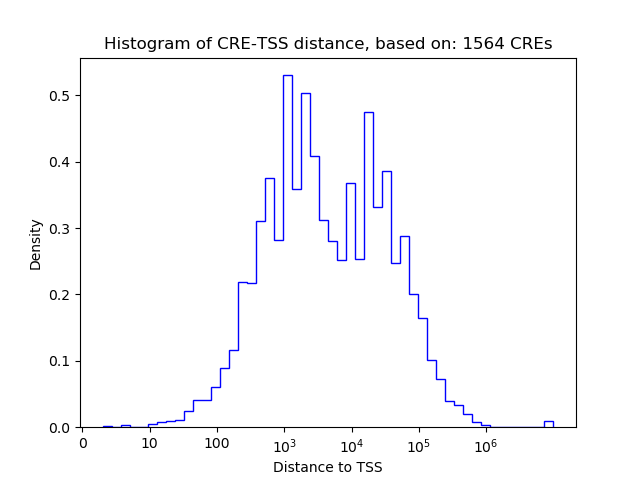
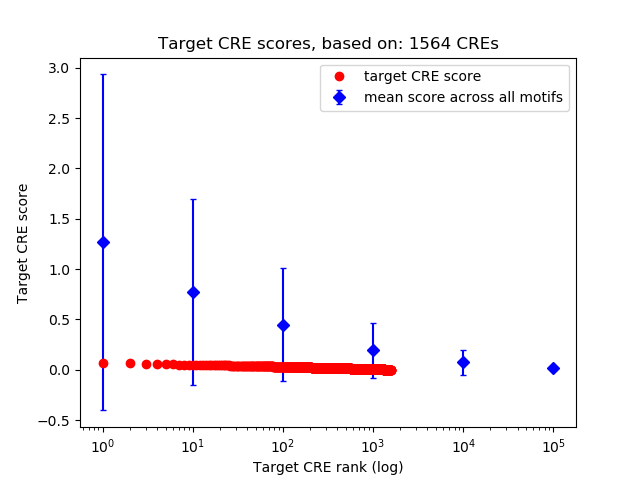
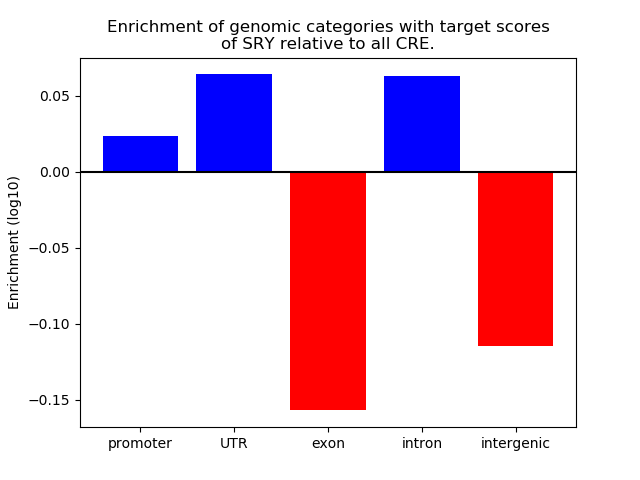
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_209930701_209930998 | 0.07 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
994 |
0.46 |
| chr7_37448490_37448719 | 0.07 |
ENSG00000200113 |
. |
10627 |
0.2 |
| chr2_204571383_204571580 | 0.06 |
CD28 |
CD28 molecule |
65 |
0.98 |
| chr13_74806186_74806399 | 0.05 |
ENSG00000206617 |
. |
57059 |
0.16 |
| chr1_160547631_160547822 | 0.05 |
CD84 |
CD84 molecule |
1537 |
0.33 |
| chrX_30594869_30596024 | 0.05 |
CXorf21 |
chromosome X open reading frame 21 |
515 |
0.84 |
| chr15_81594027_81594306 | 0.05 |
IL16 |
interleukin 16 |
2409 |
0.29 |
| chr13_31309749_31309959 | 0.05 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
209 |
0.96 |
| chr3_71542168_71542650 | 0.05 |
ENSG00000221264 |
. |
48831 |
0.15 |
| chr3_151911560_151911983 | 0.05 |
MBNL1 |
muscleblind-like splicing regulator 1 |
74058 |
0.11 |
| chr12_47607778_47608233 | 0.05 |
PCED1B |
PC-esterase domain containing 1B |
2047 |
0.36 |
| chr3_60062649_60062918 | 0.05 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
105200 |
0.08 |
| chr3_111266223_111266431 | 0.05 |
CD96 |
CD96 molecule |
5330 |
0.28 |
| chr6_143163152_143163303 | 0.05 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
5043 |
0.32 |
| chr7_150264558_150265605 | 0.05 |
GIMAP4 |
GTPase, IMAP family member 4 |
557 |
0.78 |
| chrX_48773434_48773585 | 0.05 |
PIM2 |
pim-2 oncogene |
497 |
0.64 |
| chr6_108143523_108143878 | 0.04 |
SCML4 |
sex comb on midleg-like 4 (Drosophila) |
1816 |
0.46 |
| chr7_149572287_149572438 | 0.04 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
1230 |
0.35 |
| chr3_3214293_3214763 | 0.04 |
CRBN |
cereblon |
6830 |
0.19 |
| chr13_99959249_99959863 | 0.04 |
GPR183 |
G protein-coupled receptor 183 |
103 |
0.97 |
| chr9_95728150_95728303 | 0.04 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1983 |
0.37 |
| chr2_161995820_161996335 | 0.04 |
TANK |
TRAF family member-associated NFKB activator |
2611 |
0.33 |
| chr6_26404189_26404392 | 0.04 |
BTN3A1 |
butyrophilin, subfamily 3, member A1 |
1767 |
0.21 |
| chr12_110517460_110517751 | 0.04 |
C12orf76 |
chromosome 12 open reading frame 76 |
6114 |
0.22 |
| chr5_35852941_35853144 | 0.04 |
IL7R |
interleukin 7 receptor |
245 |
0.93 |
| chr10_12408131_12408282 | 0.04 |
CAMK1D |
calcium/calmodulin-dependent protein kinase ID |
16474 |
0.18 |
| chr12_109028220_109028371 | 0.04 |
SELPLG |
selectin P ligand |
560 |
0.6 |
| chr1_111421280_111421431 | 0.04 |
CD53 |
CD53 molecule |
5579 |
0.21 |
| chr2_197032904_197033284 | 0.04 |
STK17B |
serine/threonine kinase 17b |
2630 |
0.28 |
| chr5_130882612_130883040 | 0.04 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
14100 |
0.29 |
| chr2_87799012_87799242 | 0.04 |
RP11-1399P15.1 |
|
21574 |
0.26 |
| chr10_22944496_22944670 | 0.04 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
58454 |
0.15 |
| chr7_106507057_106507208 | 0.04 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1208 |
0.59 |
| chr1_212769581_212769895 | 0.04 |
ATF3 |
activating transcription factor 3 |
12274 |
0.15 |
| chrX_135862199_135862350 | 0.04 |
ARHGEF6 |
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
1973 |
0.31 |
| chr7_137619960_137620169 | 0.04 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
1428 |
0.46 |
| chr2_225266563_225267117 | 0.04 |
FAM124B |
family with sequence similarity 124B |
38 |
0.99 |
| chr14_53593361_53593728 | 0.04 |
DDHD1 |
DDHD domain containing 1 |
23260 |
0.17 |
| chr3_151922152_151922513 | 0.04 |
MBNL1 |
muscleblind-like splicing regulator 1 |
63497 |
0.12 |
| chr13_52395252_52395592 | 0.04 |
RP11-327P2.5 |
|
16989 |
0.18 |
| chr18_72939308_72939459 | 0.04 |
TSHZ1 |
teashirt zinc finger homeobox 1 |
6370 |
0.26 |
| chr12_25207808_25208034 | 0.04 |
LRMP |
lymphoid-restricted membrane protein |
2247 |
0.32 |
| chr2_174793227_174793378 | 0.04 |
SP3 |
Sp3 transcription factor |
35645 |
0.23 |
| chr9_71396707_71396872 | 0.04 |
FAM122A |
family with sequence similarity 122A |
1825 |
0.41 |
| chr1_100820231_100820573 | 0.04 |
CDC14A |
cell division cycle 14A |
1897 |
0.33 |
| chr6_154570961_154571273 | 0.04 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
2301 |
0.45 |
| chr4_11473602_11473900 | 0.04 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
42362 |
0.18 |
| chrX_70415693_70415844 | 0.04 |
RP5-1091N2.9 |
|
2257 |
0.22 |
| chr4_199492_199643 | 0.04 |
ZNF876P |
zinc finger protein 876, pseudogene |
12872 |
0.19 |
| chr10_17726067_17726351 | 0.04 |
ENSG00000251959 |
. |
5022 |
0.18 |
| chr12_47609896_47610314 | 0.04 |
PCED1B |
PC-esterase domain containing 1B |
53 |
0.98 |
| chr2_201985519_201985852 | 0.04 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
1515 |
0.3 |
| chr17_75663137_75663644 | 0.04 |
SEPT9 |
septin 9 |
185395 |
0.03 |
| chr1_234536476_234536730 | 0.04 |
COA6 |
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
27156 |
0.13 |
| chr14_88472278_88472651 | 0.04 |
GPR65 |
G protein-coupled receptor 65 |
996 |
0.51 |
| chr2_144016827_144016978 | 0.04 |
RP11-190J23.1 |
|
87161 |
0.1 |
| chr1_231113322_231113566 | 0.04 |
TTC13 |
tetratricopeptide repeat domain 13 |
1098 |
0.34 |
| chr7_140176949_140177100 | 0.04 |
MKRN1 |
makorin ring finger protein 1 |
1279 |
0.46 |
| chr17_38021542_38021693 | 0.04 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1176 |
0.38 |
| chr4_153431598_153431794 | 0.04 |
ENSG00000264678 |
. |
21128 |
0.17 |
| chr9_3528863_3529151 | 0.03 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
3003 |
0.37 |
| chr15_31623203_31623354 | 0.03 |
KLF13 |
Kruppel-like factor 13 |
4220 |
0.35 |
| chr8_66751055_66751459 | 0.03 |
PDE7A |
phosphodiesterase 7A |
274 |
0.95 |
| chr2_197029492_197029688 | 0.03 |
STK17B |
serine/threonine kinase 17b |
6134 |
0.21 |
| chr12_15110215_15110471 | 0.03 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
3857 |
0.2 |
| chr13_41589998_41590808 | 0.03 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
3047 |
0.26 |
| chr2_158274562_158274945 | 0.03 |
CYTIP |
cytohesin 1 interacting protein |
21173 |
0.2 |
| chr12_10019991_10020525 | 0.03 |
RP11-290C10.1 |
|
632 |
0.61 |
| chr3_16929685_16929836 | 0.03 |
PLCL2 |
phospholipase C-like 2 |
3308 |
0.33 |
| chr5_158302747_158302898 | 0.03 |
CTD-2363C16.1 |
|
107192 |
0.07 |
| chr12_92531544_92532018 | 0.03 |
C12orf79 |
chromosome 12 open reading frame 79 |
984 |
0.53 |
| chr13_95923472_95923623 | 0.03 |
ABCC4 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
30042 |
0.17 |
| chr14_45719805_45719966 | 0.03 |
MIS18BP1 |
MIS18 binding protein 1 |
2495 |
0.3 |
| chr14_22594490_22594692 | 0.03 |
ENSG00000238634 |
. |
16296 |
0.27 |
| chrX_21942836_21942987 | 0.03 |
SMS |
spermine synthase |
15780 |
0.2 |
| chr3_42679974_42680125 | 0.03 |
ZBTB47 |
zinc finger and BTB domain containing 47 |
15127 |
0.1 |
| chr1_198620283_198620434 | 0.03 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
12066 |
0.23 |
| chr2_112208811_112209043 | 0.03 |
ENSG00000266139 |
. |
130259 |
0.06 |
| chr6_90789489_90789772 | 0.03 |
ENSG00000222078 |
. |
78405 |
0.1 |
| chr18_72873277_72873679 | 0.03 |
ZADH2 |
zinc binding alcohol dehydrogenase domain containing 2 |
43990 |
0.18 |
| chr1_167483086_167483237 | 0.03 |
CD247 |
CD247 molecule |
4614 |
0.23 |
| chr5_56594977_56595231 | 0.03 |
GPBP1 |
GC-rich promoter binding protein 1 |
85146 |
0.09 |
| chr4_68410033_68410229 | 0.03 |
CENPC |
centromere protein C |
1193 |
0.53 |
| chr12_56754222_56754408 | 0.03 |
STAT2 |
signal transducer and activator of transcription 2, 113kDa |
405 |
0.67 |
| chr5_139948767_139948946 | 0.03 |
SLC35A4 |
solute carrier family 35, member A4 |
4380 |
0.09 |
| chr6_2222723_2222882 | 0.03 |
GMDS |
GDP-mannose 4,6-dehydratase |
23124 |
0.28 |
| chr14_52784141_52784292 | 0.03 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
3103 |
0.33 |
| chr1_198492691_198493057 | 0.03 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
16930 |
0.28 |
| chr1_158980454_158980701 | 0.03 |
IFI16 |
interferon, gamma-inducible protein 16 |
777 |
0.67 |
| chr1_239880855_239881135 | 0.03 |
ENSG00000233355 |
. |
1371 |
0.43 |
| chr12_12874334_12874808 | 0.03 |
RP11-180M15.4 |
|
2374 |
0.2 |
| chr12_68024269_68024420 | 0.03 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
17774 |
0.25 |
| chr8_113941673_113941824 | 0.03 |
CSMD3 |
CUB and Sushi multiple domains 3 |
239477 |
0.02 |
| chr11_5246233_5246455 | 0.03 |
ENSG00000221031 |
. |
475 |
0.58 |
| chr21_36341148_36341299 | 0.03 |
RUNX1 |
runt-related transcription factor 1 |
79136 |
0.12 |
| chr19_16482667_16482930 | 0.03 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
10034 |
0.15 |
| chr6_143228096_143228637 | 0.03 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
37972 |
0.2 |
| chr13_77899440_77899640 | 0.03 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
1274 |
0.61 |
| chr6_112080304_112080455 | 0.03 |
FYN |
FYN oncogene related to SRC, FGR, YES |
45 |
0.99 |
| chr2_45794731_45794926 | 0.03 |
SRBD1 |
S1 RNA binding domain 1 |
326 |
0.88 |
| chr17_29158361_29158512 | 0.03 |
ATAD5 |
ATPase family, AAA domain containing 5 |
552 |
0.65 |
| chr2_223654006_223654270 | 0.03 |
ACSL3 |
acyl-CoA synthetase long-chain family member 3 |
71514 |
0.1 |
| chr12_118796178_118796667 | 0.03 |
TAOK3 |
TAO kinase 3 |
488 |
0.85 |
| chr10_52396469_52396620 | 0.03 |
SGMS1 |
sphingomyelin synthase 1 |
11749 |
0.18 |
| chr8_23312529_23312889 | 0.03 |
ENTPD4 |
ectonucleoside triphosphate diphosphohydrolase 4 |
2451 |
0.28 |
| chr17_7790433_7790660 | 0.03 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
1563 |
0.19 |
| chr1_109103821_109104099 | 0.03 |
FAM102B |
family with sequence similarity 102, member B |
1249 |
0.55 |
| chr12_47601705_47601886 | 0.03 |
PCED1B |
PC-esterase domain containing 1B |
8257 |
0.22 |
| chr15_75072904_75073055 | 0.03 |
CSK |
c-src tyrosine kinase |
1419 |
0.29 |
| chr6_143178538_143178701 | 0.03 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
20435 |
0.26 |
| chrY_23417410_23417561 | 0.03 |
CYorf17 |
chromosome Y open reading frame 17 |
130761 |
0.05 |
| chr7_45980495_45980766 | 0.03 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
19157 |
0.18 |
| chr10_31287543_31287794 | 0.03 |
ZNF438 |
zinc finger protein 438 |
778 |
0.75 |
| chr4_114684033_114684430 | 0.03 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
1148 |
0.66 |
| chr2_20549100_20549748 | 0.03 |
PUM2 |
pumilio RNA-binding family member 2 |
958 |
0.54 |
| chr5_39177582_39177762 | 0.03 |
FYB |
FYN binding protein |
25457 |
0.23 |
| chr3_159628787_159629360 | 0.03 |
SCHIP1 |
schwannomin interacting protein 1 |
58345 |
0.11 |
| chrX_78620075_78620267 | 0.03 |
ITM2A |
integral membrane protein 2A |
2685 |
0.44 |
| chr12_3982064_3982233 | 0.03 |
PARP11 |
poly (ADP-ribose) polymerase family, member 11 |
373 |
0.82 |
| chr14_22977314_22977663 | 0.03 |
TRAJ15 |
T cell receptor alpha joining 15 |
21092 |
0.09 |
| chr5_67515061_67515212 | 0.03 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
3532 |
0.3 |
| chr19_42390602_42391041 | 0.03 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
2306 |
0.19 |
| chr2_204803308_204803520 | 0.03 |
ICOS |
inducible T-cell co-stimulator |
1911 |
0.46 |
| chr6_159074554_159074705 | 0.03 |
SYTL3 |
synaptotagmin-like 3 |
3583 |
0.22 |
| chr15_81586254_81586487 | 0.03 |
IL16 |
interleukin 16 |
2884 |
0.28 |
| chr12_104888025_104888586 | 0.03 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
37526 |
0.2 |
| chr14_71110053_71110301 | 0.03 |
TTC9 |
tetratricopeptide repeat domain 9 |
1673 |
0.4 |
| chr10_76728189_76728340 | 0.03 |
RP11-77G23.5 |
|
55745 |
0.12 |
| chr14_91524676_91524827 | 0.03 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
1727 |
0.3 |
| chr7_5694530_5694681 | 0.03 |
RNF216-IT1 |
RNF216 intronic transcript 1 (non-protein coding) |
25487 |
0.16 |
| chrX_149739512_149739787 | 0.03 |
MTM1 |
myotubularin 1 |
2556 |
0.37 |
| chr4_71860383_71860569 | 0.03 |
DCK |
deoxycytidine kinase |
1121 |
0.6 |
| chr10_70748146_70748533 | 0.03 |
KIAA1279 |
KIAA1279 |
148 |
0.95 |
| chr5_56113673_56113824 | 0.03 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
2347 |
0.29 |
| chr12_11804572_11805680 | 0.03 |
ETV6 |
ets variant 6 |
2338 |
0.4 |
| chr4_40205321_40205527 | 0.03 |
RHOH |
ras homolog family member H |
3460 |
0.27 |
| chr16_87889915_87890443 | 0.03 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
7066 |
0.18 |
| chr5_137911540_137911767 | 0.03 |
HSPA9 |
heat shock 70kDa protein 9 (mortalin) |
520 |
0.71 |
| chr14_22994017_22994177 | 0.03 |
TRAJ15 |
T cell receptor alpha joining 15 |
4483 |
0.13 |
| chr2_55236798_55237891 | 0.03 |
RTN4 |
reticulon 4 |
241 |
0.93 |
| chr19_42038250_42038705 | 0.03 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
17409 |
0.15 |
| chr2_109239891_109240042 | 0.03 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
2244 |
0.37 |
| chr12_15716958_15717148 | 0.03 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
16644 |
0.26 |
| chr2_231921398_231921578 | 0.03 |
PSMD1 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
90 |
0.96 |
| chr22_38793751_38794050 | 0.03 |
CSNK1E |
casein kinase 1, epsilon |
627 |
0.62 |
| chr14_38558222_38558398 | 0.03 |
CTD-2058B24.2 |
|
2053 |
0.43 |
| chr16_22383541_22383722 | 0.03 |
CDR2 |
cerebellar degeneration-related protein 2, 62kDa |
2307 |
0.21 |
| chr16_71851540_71851781 | 0.03 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
8556 |
0.13 |
| chr16_84793452_84793603 | 0.03 |
USP10 |
ubiquitin specific peptidase 10 |
8344 |
0.22 |
| chr19_12061488_12061639 | 0.03 |
ZNF700 |
zinc finger protein 700 |
3994 |
0.13 |
| chr7_150374007_150374169 | 0.03 |
GIMAP2 |
GTPase, IMAP family member 2 |
8700 |
0.17 |
| chr1_52014581_52014732 | 0.03 |
ENSG00000252032 |
. |
10255 |
0.16 |
| chrX_74376165_74376390 | 0.03 |
ABCB7 |
ATP-binding cassette, sub-family B (MDR/TAP), member 7 |
145 |
0.97 |
| chr16_46722355_46722638 | 0.03 |
VPS35 |
vacuolar protein sorting 35 homolog (S. cerevisiae) |
608 |
0.58 |
| chr5_37198039_37198190 | 0.03 |
C5orf42 |
chromosome 5 open reading frame 42 |
15610 |
0.24 |
| chr7_28369849_28370167 | 0.03 |
CREB5 |
cAMP responsive element binding protein 5 |
31068 |
0.23 |
| chr15_86125606_86125830 | 0.03 |
RP11-815J21.2 |
|
2309 |
0.27 |
| chr8_8706094_8706267 | 0.03 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
44975 |
0.14 |
| chr12_11805836_11806191 | 0.03 |
ETV6 |
ets variant 6 |
3225 |
0.34 |
| chr2_43448588_43448818 | 0.03 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
5045 |
0.26 |
| chr2_198169566_198169717 | 0.03 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
2398 |
0.25 |
| chrX_78417033_78417184 | 0.03 |
GPR174 |
G protein-coupled receptor 174 |
9361 |
0.32 |
| chr8_17554451_17554629 | 0.03 |
MTUS1 |
microtubule associated tumor suppressor 1 |
639 |
0.69 |
| chr10_14604221_14604372 | 0.03 |
FAM107B |
family with sequence similarity 107, member B |
6117 |
0.27 |
| chr2_202127394_202127951 | 0.02 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
993 |
0.56 |
| chr13_41557045_41557196 | 0.02 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
702 |
0.71 |
| chr3_107319287_107319503 | 0.02 |
BBX |
bobby sox homolog (Drosophila) |
1235 |
0.65 |
| chr9_32525867_32526215 | 0.02 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
152 |
0.95 |
| chr4_76598858_76599149 | 0.02 |
G3BP2 |
GTPase activating protein (SH3 domain) binding protein 2 |
150 |
0.95 |
| chr3_151989912_151990063 | 0.02 |
MBNL1-AS1 |
MBNL1 antisense RNA 1 |
2643 |
0.28 |
| chr8_41649611_41650018 | 0.02 |
ANK1 |
ankyrin 1, erythrocytic |
5326 |
0.2 |
| chr4_80886303_80886454 | 0.02 |
ANTXR2 |
anthrax toxin receptor 2 |
107339 |
0.07 |
| chr2_198102391_198102542 | 0.02 |
ANKRD44 |
ankyrin repeat domain 44 |
39704 |
0.14 |
| chr2_37844911_37845062 | 0.02 |
AC006369.2 |
|
17707 |
0.22 |
| chr4_38857928_38858431 | 0.02 |
TLR6 |
toll-like receptor 6 |
258 |
0.48 |
| chr9_35276767_35276918 | 0.02 |
AL160274.1 |
HCG17281; PRO0038; Uncharacterized protein |
84420 |
0.07 |
| chr13_48809687_48810120 | 0.02 |
ITM2B |
integral membrane protein 2B |
2564 |
0.37 |
| chr5_64447748_64447899 | 0.02 |
ENSG00000207439 |
. |
28627 |
0.26 |
| chr6_152622546_152623206 | 0.02 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
382 |
0.91 |
| chr21_36413445_36413809 | 0.02 |
RUNX1 |
runt-related transcription factor 1 |
7835 |
0.33 |
| chr6_150739556_150740117 | 0.02 |
IYD |
iodotyrosine deiodinase |
48723 |
0.16 |
| chrX_153626756_153627188 | 0.02 |
RPL10 |
ribosomal protein L10 |
253 |
0.78 |
| chr21_48025776_48026061 | 0.02 |
S100B |
S100 calcium binding protein B |
797 |
0.66 |
| chr1_25663249_25663400 | 0.02 |
TMEM50A |
transmembrane protein 50A |
1084 |
0.32 |
| chr2_87883698_87883984 | 0.02 |
ENSG00000265507 |
. |
45433 |
0.18 |
| chr7_15214941_15215142 | 0.02 |
AGMO |
alkylglycerol monooxygenase |
190771 |
0.03 |
| chr2_214148558_214149019 | 0.02 |
SPAG16 |
sperm associated antigen 16 |
325 |
0.94 |
| chr1_230201927_230202153 | 0.02 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
916 |
0.66 |
| chr5_32171141_32171315 | 0.02 |
GOLPH3 |
golgi phosphoprotein 3 (coat-protein) |
3228 |
0.25 |
| chr15_86098904_86099149 | 0.02 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
349 |
0.88 |
| chr12_15104822_15105163 | 0.02 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
907 |
0.54 |
| chr19_16493261_16493427 | 0.02 |
ENSG00000243745 |
. |
16426 |
0.13 |
| chr9_94877804_94877985 | 0.02 |
SPTLC1 |
serine palmitoyltransferase, long chain base subunit 1 |
229 |
0.94 |
| chr11_73690068_73690219 | 0.02 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
957 |
0.48 |
| chr5_131310633_131310784 | 0.02 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
19210 |
0.13 |
| chr12_62656333_62656484 | 0.02 |
USP15 |
ubiquitin specific peptidase 15 |
2199 |
0.3 |
| chr1_102637571_102637722 | 0.02 |
OLFM3 |
olfactomedin 3 |
175060 |
0.04 |
| chr12_32113022_32113222 | 0.02 |
KIAA1551 |
KIAA1551 |
769 |
0.71 |
| chr19_17493496_17493698 | 0.02 |
PLVAP |
plasmalemma vesicle associated protein |
5438 |
0.09 |
| chr17_33400509_33400660 | 0.02 |
RFFL |
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
9713 |
0.1 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |