Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for STAT1_STAT3_BCL6
Z-value: 0.50
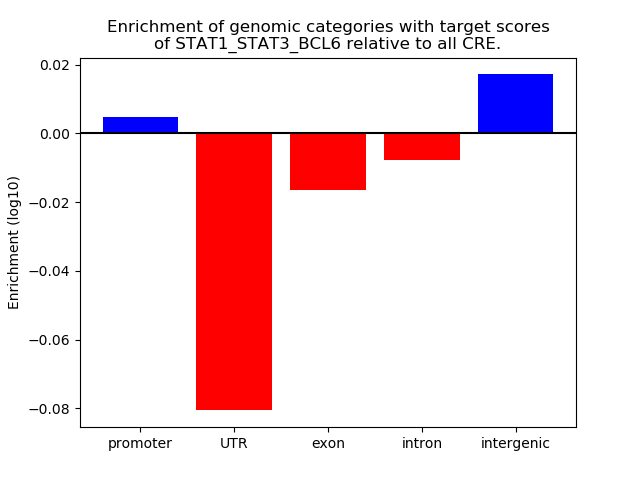
Transcription factors associated with STAT1_STAT3_BCL6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
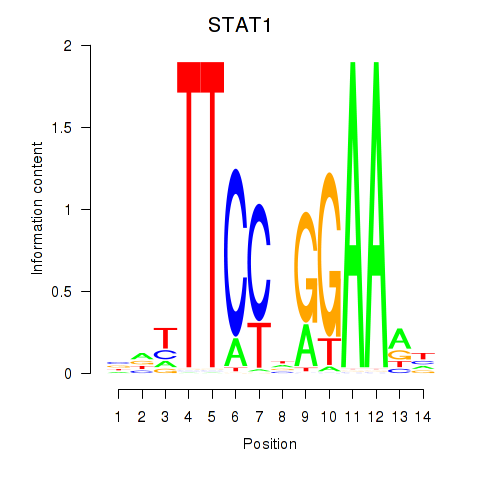
STAT1
|
ENSG00000115415.14 | signal transducer and activator of transcription 1 |
|
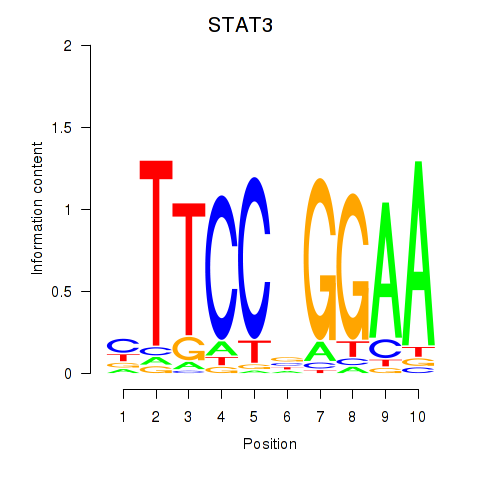
STAT3
|
ENSG00000168610.10 | signal transducer and activator of transcription 3 |
|
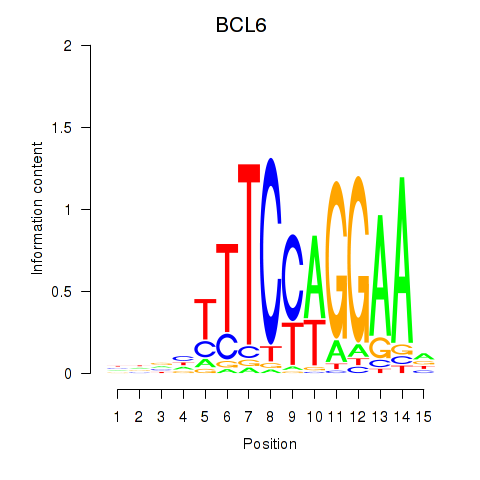
BCL6
|
ENSG00000113916.13 | BCL6 transcription repressor |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr3_187520878_187521148 | BCL6 | 57498 | 0.139536 | -0.86 | 2.9e-03 | Click! |
| chr3_187456856_187457007 | BCL6 | 1199 | 0.515166 | -0.83 | 5.1e-03 | Click! |
| chr3_187485668_187485819 | BCL6 | 22228 | 0.213382 | 0.81 | 7.7e-03 | Click! |
| chr3_187637243_187637630 | BCL6 | 173921 | 0.031913 | -0.77 | 1.5e-02 | Click! |
| chr3_187454096_187454438 | BCL6 | 90 | 0.974123 | 0.73 | 2.6e-02 | Click! |
| chr2_191876732_191876887 | STAT1 | 1504 | 0.371062 | -0.87 | 2.4e-03 | Click! |
| chr2_191877453_191877604 | STAT1 | 785 | 0.612427 | -0.87 | 2.5e-03 | Click! |
| chr2_191877633_191877798 | STAT1 | 598 | 0.715316 | -0.85 | 3.7e-03 | Click! |
| chr2_191877894_191878160 | STAT1 | 286 | 0.895648 | -0.77 | 1.5e-02 | Click! |
| chr2_191876019_191876170 | STAT1 | 2219 | 0.276694 | -0.66 | 5.1e-02 | Click! |
| chr17_40507445_40507596 | STAT3 | 32912 | 0.094354 | 0.73 | 2.6e-02 | Click! |
| chr17_40516095_40516246 | STAT3 | 24262 | 0.106798 | 0.73 | 2.7e-02 | Click! |
| chr17_40540908_40541151 | STAT3 | 443 | 0.738460 | -0.52 | 1.5e-01 | Click! |
| chr17_40534451_40534602 | STAT3 | 5906 | 0.131894 | 0.51 | 1.6e-01 | Click! |
| chr17_40535679_40535830 | STAT3 | 4678 | 0.139398 | 0.45 | 2.2e-01 | Click! |
Activity of the STAT1_STAT3_BCL6 motif across conditions
Conditions sorted by the z-value of the STAT1_STAT3_BCL6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
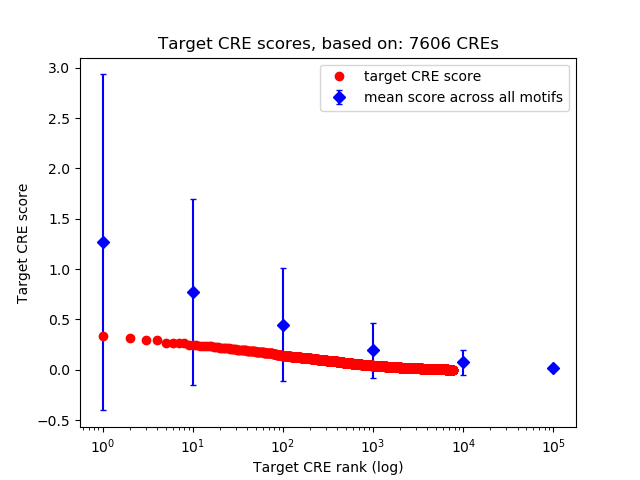
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr8_104384541_104384884 | 0.34 |
CTHRC1 |
collagen triple helix repeat containing 1 |
51 |
0.97 |
| chr2_66802213_66802500 | 0.32 |
MEIS1 |
Meis homeobox 1 |
66297 |
0.13 |
| chrX_67913478_67914353 | 0.30 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
422 |
0.91 |
| chr9_124320076_124320249 | 0.30 |
DAB2IP |
DAB2 interacting protein |
9174 |
0.18 |
| chr12_2163640_2163867 | 0.27 |
CACNA1C |
calcium channel, voltage-dependent, L type, alpha 1C subunit |
1024 |
0.49 |
| chr12_53443889_53444561 | 0.27 |
TENC1 |
tensin like C1 domain containing phosphatase (tensin 2) |
255 |
0.86 |
| chr5_15620667_15620944 | 0.27 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
4714 |
0.28 |
| chrX_34673666_34673817 | 0.27 |
TMEM47 |
transmembrane protein 47 |
1664 |
0.56 |
| chr15_63337269_63337696 | 0.25 |
TPM1 |
tropomyosin 1 (alpha) |
1507 |
0.37 |
| chrX_114467670_114468385 | 0.24 |
LRCH2 |
leucine-rich repeats and calponin homology (CH) domain containing 2 |
601 |
0.77 |
| chr3_145968965_145969195 | 0.24 |
PLSCR4 |
phospholipid scramblase 4 |
114 |
0.98 |
| chr14_73929626_73929971 | 0.24 |
ENSG00000251393 |
. |
669 |
0.64 |
| chr1_186649638_186650434 | 0.23 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
477 |
0.89 |
| chr14_55239224_55239463 | 0.23 |
SAMD4A |
sterile alpha motif domain containing 4A |
17792 |
0.24 |
| chr7_38624277_38624485 | 0.23 |
AMPH |
amphiphysin |
46639 |
0.16 |
| chr2_38303069_38303895 | 0.23 |
CYP1B1-AS1 |
CYP1B1 antisense RNA 1 |
156 |
0.5 |
| chr5_33891044_33891453 | 0.23 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
798 |
0.63 |
| chr17_42214986_42215221 | 0.22 |
ENSG00000212446 |
. |
1746 |
0.18 |
| chr17_38096107_38096317 | 0.22 |
LRRC3C |
leucine rich repeat containing 3C |
1515 |
0.27 |
| chr1_162472131_162472282 | 0.22 |
UHMK1 |
U2AF homology motif (UHM) kinase 1 |
4573 |
0.22 |
| chr7_100200266_100200615 | 0.22 |
PCOLCE-AS1 |
PCOLCE antisense RNA 1 |
218 |
0.74 |
| chr3_55519561_55519953 | 0.22 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1574 |
0.4 |
| chr2_14775112_14775752 | 0.21 |
AC011897.1 |
Uncharacterized protein |
217 |
0.95 |
| chr10_42862871_42863499 | 0.21 |
RP11-313J2.1 |
|
14726 |
0.27 |
| chr15_48960228_48960461 | 0.21 |
FBN1 |
fibrillin 1 |
22298 |
0.23 |
| chr22_24721245_24721396 | 0.21 |
SPECC1L |
sperm antigen with calponin homology and coiled-coil domains 1-like |
20956 |
0.18 |
| chr6_125285262_125285420 | 0.20 |
RP11-510H23.1 |
|
1156 |
0.45 |
| chr16_3432471_3432622 | 0.20 |
MTRNR2L4 |
MT-RNR2-like 4 |
10263 |
0.11 |
| chr6_16318510_16318713 | 0.20 |
GMPR |
guanosine monophosphate reductase |
79800 |
0.09 |
| chr13_32520909_32521060 | 0.20 |
EEF1DP3 |
eukaryotic translation elongation factor 1 delta pseudogene 3 |
5733 |
0.3 |
| chr4_170188382_170188561 | 0.20 |
SH3RF1 |
SH3 domain containing ring finger 1 |
2637 |
0.38 |
| chr8_136471145_136471318 | 0.20 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
281 |
0.92 |
| chr5_167696137_167696498 | 0.20 |
WWC1 |
WW and C2 domain containing 1 |
22339 |
0.2 |
| chr7_132299406_132299928 | 0.20 |
PLXNA4 |
plexin A4 |
33780 |
0.18 |
| chr2_218874347_218874698 | 0.20 |
TNS1 |
tensin 1 |
6804 |
0.19 |
| chr4_175750179_175750687 | 0.19 |
GLRA3 |
glycine receptor, alpha 3 |
32 |
0.99 |
| chr6_3749707_3750198 | 0.19 |
RP11-420L9.5 |
|
1393 |
0.41 |
| chr12_56138916_56139328 | 0.19 |
GDF11 |
growth differentiation factor 11 |
1939 |
0.16 |
| chr12_77625749_77626115 | 0.19 |
ENSG00000238769 |
. |
69127 |
0.14 |
| chr10_91055374_91055594 | 0.19 |
IFIT2 |
interferon-induced protein with tetratricopeptide repeats 2 |
6228 |
0.15 |
| chr9_123698545_123698822 | 0.19 |
TRAF1 |
TNF receptor-associated factor 1 |
7232 |
0.23 |
| chr12_81471441_81472090 | 0.19 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
71 |
0.98 |
| chr19_55684291_55685412 | 0.19 |
SYT5 |
synaptotagmin V |
1083 |
0.28 |
| chr2_223163063_223163341 | 0.19 |
PAX3 |
paired box 3 |
263 |
0.57 |
| chr3_136558029_136558180 | 0.18 |
RP11-731C17.2 |
|
241 |
0.91 |
| chr2_159825200_159826543 | 0.18 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
688 |
0.73 |
| chr9_21764802_21765039 | 0.18 |
MTAP |
methylthioadenosine phosphorylase |
37622 |
0.15 |
| chr3_123602332_123603013 | 0.18 |
MYLK |
myosin light chain kinase |
477 |
0.85 |
| chr5_82768204_82768925 | 0.18 |
VCAN |
versican |
820 |
0.75 |
| chr5_150181351_150181502 | 0.18 |
AC010441.1 |
|
23559 |
0.14 |
| chr15_39871853_39872515 | 0.18 |
THBS1 |
thrombospondin 1 |
1096 |
0.52 |
| chr10_121529608_121529837 | 0.18 |
ENSG00000242818 |
. |
1963 |
0.36 |
| chr15_50352056_50352408 | 0.18 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
12648 |
0.24 |
| chr11_65639318_65639911 | 0.18 |
EFEMP2 |
EGF containing fibulin-like extracellular matrix protein 2 |
511 |
0.56 |
| chr18_34275433_34275584 | 0.18 |
FHOD3 |
formin homology 2 domain containing 3 |
23012 |
0.23 |
| chr7_134540880_134541057 | 0.18 |
CALD1 |
caldesmon 1 |
10624 |
0.29 |
| chr5_65893484_65893635 | 0.18 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
1075 |
0.66 |
| chr17_21452935_21453464 | 0.18 |
C17orf51 |
chromosome 17 open reading frame 51 |
1744 |
0.38 |
| chr17_57504883_57505034 | 0.17 |
RP11-567L7.5 |
|
24294 |
0.18 |
| chr12_8090559_8091101 | 0.17 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
1959 |
0.27 |
| chr8_101555325_101555476 | 0.17 |
ANKRD46 |
ankyrin repeat domain 46 |
9847 |
0.18 |
| chr4_142003365_142003602 | 0.17 |
RNF150 |
ring finger protein 150 |
28603 |
0.25 |
| chr10_28287180_28287375 | 0.17 |
ARMC4 |
armadillo repeat containing 4 |
700 |
0.81 |
| chr20_31063509_31063841 | 0.17 |
C20orf112 |
chromosome 20 open reading frame 112 |
7599 |
0.19 |
| chr2_149894818_149895436 | 0.17 |
LYPD6B |
LY6/PLAUR domain containing 6B |
105 |
0.98 |
| chr3_122478078_122478289 | 0.17 |
ENSG00000238480 |
. |
8964 |
0.19 |
| chr2_176931780_176932252 | 0.17 |
EVX2 |
even-skipped homeobox 2 |
16625 |
0.1 |
| chr10_113595251_113595472 | 0.17 |
GPAM |
glycerol-3-phosphate acyltransferase, mitochondrial |
348107 |
0.01 |
| chr15_56206351_56206878 | 0.17 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
1238 |
0.55 |
| chr3_159484310_159484461 | 0.16 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
1493 |
0.34 |
| chr2_133104359_133104833 | 0.16 |
ENSG00000252705 |
. |
59835 |
0.11 |
| chr7_94034050_94034201 | 0.16 |
COL1A2 |
collagen, type I, alpha 2 |
10252 |
0.27 |
| chr18_11983196_11983347 | 0.16 |
IMPA2 |
inositol(myo)-1(or 4)-monophosphatase 2 |
1602 |
0.34 |
| chr2_238490360_238490511 | 0.16 |
RAB17 |
RAB17, member RAS oncogene family |
4307 |
0.16 |
| chr16_50236729_50236952 | 0.16 |
ADCY7 |
adenylate cyclase 7 |
43208 |
0.12 |
| chr3_65582462_65582613 | 0.16 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
873 |
0.76 |
| chr2_145266428_145266579 | 0.16 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
8612 |
0.25 |
| chr15_83620716_83621316 | 0.16 |
HOMER2 |
homer homolog 2 (Drosophila) |
457 |
0.76 |
| chr3_75690482_75691208 | 0.16 |
ENSG00000221795 |
. |
10931 |
0.2 |
| chr1_90158313_90158464 | 0.16 |
LRRC8C |
leucine rich repeat containing 8 family, member C |
59757 |
0.1 |
| chr22_50698815_50699396 | 0.16 |
MAPK12 |
mitogen-activated protein kinase 12 |
632 |
0.49 |
| chr1_205605987_205606138 | 0.16 |
ELK4 |
ELK4, ETS-domain protein (SRF accessory protein 1) |
4972 |
0.18 |
| chr12_12111873_12112098 | 0.16 |
ETV6 |
ets variant 6 |
73114 |
0.11 |
| chr3_189794073_189794333 | 0.15 |
ENSG00000265045 |
. |
37520 |
0.14 |
| chr2_175830663_175830896 | 0.15 |
ENSG00000201425 |
. |
11752 |
0.19 |
| chrX_11446077_11446283 | 0.15 |
ARHGAP6 |
Rho GTPase activating protein 6 |
287 |
0.95 |
| chr15_38693567_38693718 | 0.15 |
FAM98B |
family with sequence similarity 98, member B |
52686 |
0.15 |
| chr1_225839301_225840394 | 0.15 |
ENAH |
enabled homolog (Drosophila) |
571 |
0.78 |
| chrX_106045541_106045714 | 0.15 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
283 |
0.94 |
| chr10_121209563_121209788 | 0.15 |
GRK5 |
G protein-coupled receptor kinase 5 |
71726 |
0.09 |
| chr7_19155354_19155672 | 0.15 |
TWIST1 |
twist family bHLH transcription factor 1 |
1782 |
0.28 |
| chr4_166252557_166253372 | 0.15 |
MSMO1 |
methylsterol monooxygenase 1 |
3885 |
0.25 |
| chr15_64231749_64232239 | 0.15 |
RP11-111E14.1 |
|
11521 |
0.22 |
| chr1_39284364_39284540 | 0.14 |
RRAGC |
Ras-related GTP binding C |
41043 |
0.12 |
| chr16_87096172_87096484 | 0.14 |
RP11-899L11.3 |
|
153193 |
0.04 |
| chr11_93582978_93583471 | 0.14 |
VSTM5 |
V-set and transmembrane domain containing 5 |
473 |
0.74 |
| chr18_46581903_46582348 | 0.14 |
ENSG00000263849 |
. |
5987 |
0.21 |
| chr1_94158748_94158940 | 0.14 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
11459 |
0.24 |
| chr20_49307514_49308405 | 0.14 |
FAM65C |
family with sequence similarity 65, member C |
43 |
0.97 |
| chr3_149094469_149095370 | 0.14 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
646 |
0.47 |
| chr14_59331604_59331755 | 0.14 |
ENSG00000221427 |
. |
93715 |
0.09 |
| chr18_65983753_65984295 | 0.14 |
TMX3 |
thioredoxin-related transmembrane protein 3 |
398270 |
0.01 |
| chr2_85361754_85362037 | 0.14 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
1362 |
0.47 |
| chr7_50859466_50860386 | 0.14 |
GRB10 |
growth factor receptor-bound protein 10 |
740 |
0.77 |
| chr18_60052258_60052436 | 0.14 |
RP11-640A1.3 |
|
6984 |
0.22 |
| chr5_158488269_158488591 | 0.14 |
EBF1 |
early B-cell factor 1 |
38271 |
0.17 |
| chr16_14458625_14458905 | 0.14 |
ENSG00000199130 |
. |
55623 |
0.12 |
| chr10_3828809_3828960 | 0.14 |
KLF6 |
Kruppel-like factor 6 |
1411 |
0.47 |
| chr1_203247491_203247930 | 0.14 |
BTG2 |
BTG family, member 2 |
26954 |
0.14 |
| chr6_164404752_164405006 | 0.14 |
ENSG00000266128 |
. |
143286 |
0.05 |
| chr10_115860454_115861476 | 0.14 |
ENSG00000253066 |
. |
10208 |
0.2 |
| chr3_170806844_170806995 | 0.14 |
ENSG00000207963 |
. |
17629 |
0.23 |
| chr1_119543090_119543475 | 0.14 |
TBX15 |
T-box 15 |
11103 |
0.25 |
| chr16_87250830_87251124 | 0.13 |
RP11-899L11.3 |
|
1456 |
0.45 |
| chr8_23398759_23399053 | 0.13 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
12333 |
0.16 |
| chr15_96899977_96900177 | 0.13 |
AC087477.1 |
Uncharacterized protein |
4410 |
0.2 |
| chr22_47330549_47330700 | 0.13 |
ENSG00000221672 |
. |
86821 |
0.1 |
| chr14_50413720_50413871 | 0.13 |
ENSG00000251929 |
. |
45127 |
0.1 |
| chr12_30353916_30354159 | 0.13 |
ENSG00000251781 |
. |
105420 |
0.08 |
| chr12_113859774_113860410 | 0.13 |
SDSL |
serine dehydratase-like |
50 |
0.97 |
| chr14_102604980_102605131 | 0.13 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
968 |
0.41 |
| chr18_52626293_52626952 | 0.13 |
CCDC68 |
coiled-coil domain containing 68 |
117 |
0.97 |
| chr16_3341320_3341471 | 0.13 |
ZNF263 |
zinc finger protein 263 |
992 |
0.37 |
| chr4_184492781_184492932 | 0.13 |
ENSG00000239116 |
. |
10598 |
0.19 |
| chr6_32489639_32489790 | 0.13 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
8350 |
0.15 |
| chr7_87936252_87936498 | 0.13 |
STEAP4 |
STEAP family member 4 |
169 |
0.96 |
| chr5_132112834_132113698 | 0.13 |
SEPT8 |
septin 8 |
153 |
0.93 |
| chr4_8594931_8595082 | 0.13 |
CPZ |
carboxypeptidase Z |
523 |
0.82 |
| chr10_103330110_103330718 | 0.13 |
DPCD |
deleted in primary ciliary dyskinesia homolog (mouse) |
8 |
0.98 |
| chr12_88973452_88974158 | 0.13 |
KITLG |
KIT ligand |
433 |
0.82 |
| chr11_89113858_89114276 | 0.13 |
NOX4 |
NADPH oxidase 4 |
109833 |
0.07 |
| chr8_125827427_125827626 | 0.13 |
ENSG00000263735 |
. |
6774 |
0.28 |
| chr8_99304651_99304929 | 0.13 |
NIPAL2 |
NIPA-like domain containing 2 |
1774 |
0.45 |
| chr3_128268180_128268331 | 0.13 |
C3orf27 |
chromosome 3 open reading frame 27 |
26674 |
0.16 |
| chr10_29980674_29981313 | 0.13 |
ENSG00000222092 |
. |
19851 |
0.22 |
| chr3_71294181_71294421 | 0.13 |
FOXP1 |
forkhead box P1 |
15 |
0.99 |
| chr17_16159063_16159236 | 0.13 |
ENSG00000221355 |
. |
26179 |
0.13 |
| chr4_55098050_55098472 | 0.13 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
1772 |
0.47 |
| chr19_30941172_30941421 | 0.13 |
ZNF536 |
zinc finger protein 536 |
77977 |
0.12 |
| chr15_75018290_75019298 | 0.13 |
CYP1A1 |
cytochrome P450, family 1, subfamily A, polypeptide 1 |
843 |
0.52 |
| chr17_13444026_13444182 | 0.13 |
ENSG00000221698 |
. |
2859 |
0.3 |
| chr16_59669437_59669588 | 0.13 |
ENSG00000200062 |
. |
95482 |
0.09 |
| chr13_27557206_27557734 | 0.13 |
USP12-AS1 |
USP12 antisense RNA 1 |
179522 |
0.03 |
| chr22_48594670_48594821 | 0.13 |
ENSG00000266508 |
. |
75431 |
0.13 |
| chr11_61448104_61448359 | 0.12 |
DAGLA |
diacylglycerol lipase, alpha |
326 |
0.89 |
| chr8_42358295_42358694 | 0.12 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
267 |
0.92 |
| chr7_27143412_27143563 | 0.12 |
HOXA2 |
homeobox A2 |
1057 |
0.27 |
| chr5_151065704_151066114 | 0.12 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
615 |
0.66 |
| chr21_17567832_17567983 | 0.12 |
ENSG00000201025 |
. |
89182 |
0.1 |
| chr10_101754409_101754592 | 0.12 |
DNMBP |
dynamin binding protein |
15176 |
0.18 |
| chr2_173037089_173037327 | 0.12 |
ENSG00000238572 |
. |
16360 |
0.23 |
| chr5_52105955_52106201 | 0.12 |
CTD-2288O8.1 |
|
22218 |
0.18 |
| chr1_1934983_1935264 | 0.12 |
C1orf222 |
chromosome 1 open reading frame 222 |
30 |
0.96 |
| chr22_45403768_45404420 | 0.12 |
PHF21B |
PHD finger protein 21B |
736 |
0.7 |
| chr17_32582009_32582182 | 0.12 |
CCL2 |
chemokine (C-C motif) ligand 2 |
209 |
0.48 |
| chr15_67840467_67840618 | 0.12 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
794 |
0.7 |
| chr18_43304377_43304558 | 0.12 |
SLC14A1 |
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
318 |
0.9 |
| chr14_33104283_33104434 | 0.12 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
100699 |
0.08 |
| chr6_152084748_152085135 | 0.12 |
ESR1 |
estrogen receptor 1 |
40625 |
0.17 |
| chr1_185825882_185826255 | 0.12 |
HMCN1 |
hemicentin 1 |
122385 |
0.06 |
| chr9_93823154_93823334 | 0.12 |
SYK |
spleen tyrosine kinase |
233474 |
0.02 |
| chr5_179757668_179757864 | 0.12 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
882 |
0.61 |
| chr12_15781229_15781461 | 0.12 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
33863 |
0.17 |
| chr6_132722750_132722944 | 0.12 |
MOXD1 |
monooxygenase, DBH-like 1 |
163 |
0.97 |
| chr1_94219763_94219914 | 0.12 |
RP11-488P3.1 |
|
21162 |
0.21 |
| chr3_167580405_167580688 | 0.12 |
SERPINI1 |
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
55520 |
0.16 |
| chr1_234669487_234669677 | 0.12 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
54733 |
0.11 |
| chr1_85557311_85557759 | 0.12 |
WDR63 |
WD repeat domain 63 |
29525 |
0.15 |
| chr16_55513844_55514378 | 0.12 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
201 |
0.95 |
| chr1_210412971_210413268 | 0.12 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
5727 |
0.24 |
| chr21_38580312_38580506 | 0.12 |
ENSG00000263969 |
. |
7497 |
0.16 |
| chr13_30983970_30984184 | 0.12 |
HMGB1 |
high mobility group box 1 |
54307 |
0.15 |
| chr4_88894663_88894869 | 0.12 |
SPP1 |
secreted phosphoprotein 1 |
2053 |
0.33 |
| chr17_77015960_77016131 | 0.12 |
C1QTNF1 |
C1q and tumor necrosis factor related protein 1 |
2851 |
0.19 |
| chr11_105482279_105482430 | 0.12 |
GRIA4 |
glutamate receptor, ionotropic, AMPA 4 |
629 |
0.84 |
| chr4_138449879_138450179 | 0.12 |
PCDH18 |
protocadherin 18 |
3536 |
0.4 |
| chr7_130594902_130595108 | 0.12 |
ENSG00000226380 |
. |
32707 |
0.2 |
| chr10_97723067_97723218 | 0.12 |
CC2D2B |
coiled-coil and C2 domain containing 2B |
10644 |
0.17 |
| chr2_238321930_238322164 | 0.12 |
COL6A3 |
collagen, type VI, alpha 3 |
744 |
0.69 |
| chr3_157156592_157156743 | 0.12 |
PTX3 |
pentraxin 3, long |
2089 |
0.41 |
| chr12_90484641_90484914 | 0.12 |
ENSG00000252823 |
. |
336941 |
0.01 |
| chr6_86162562_86162713 | 0.12 |
NT5E |
5'-nucleotidase, ecto (CD73) |
2810 |
0.37 |
| chr7_152403601_152404275 | 0.12 |
XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
30688 |
0.14 |
| chr1_149287790_149287941 | 0.12 |
ENSG00000252105 |
. |
1543 |
0.36 |
| chr8_75897244_75897545 | 0.12 |
CRISPLD1 |
cysteine-rich secretory protein LCCL domain containing 1 |
531 |
0.84 |
| chr11_65790225_65790471 | 0.12 |
CATSPER1 |
cation channel, sperm associated 1 |
3640 |
0.11 |
| chr6_113700515_113700666 | 0.12 |
ENSG00000222677 |
. |
135599 |
0.05 |
| chr12_50608832_50609096 | 0.12 |
RP3-405J10.4 |
|
4423 |
0.14 |
| chr2_107501882_107502033 | 0.12 |
ST6GAL2 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
512 |
0.85 |
| chr1_234792376_234792776 | 0.12 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
47305 |
0.14 |
| chr4_88480802_88480953 | 0.12 |
SPARCL1 |
SPARC-like 1 (hevin) |
28664 |
0.17 |
| chr14_88596087_88596243 | 0.11 |
RP11-300J18.2 |
|
114210 |
0.06 |
| chr14_78073975_78074126 | 0.11 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
9066 |
0.2 |
| chr2_151342010_151342200 | 0.11 |
RND3 |
Rho family GTPase 3 |
209 |
0.97 |
| chr5_53830476_53830627 | 0.11 |
SNX18 |
sorting nexin 18 |
16958 |
0.27 |
| chr12_16039681_16039832 | 0.11 |
STRAP |
serine/threonine kinase receptor associated protein |
4427 |
0.23 |
| chr3_114341689_114341840 | 0.11 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
1289 |
0.61 |
| chr10_30138566_30138997 | 0.11 |
SVIL |
supervillin |
114048 |
0.06 |
| chr2_205587293_205587444 | 0.11 |
PARD3B |
par-3 family cell polarity regulator beta |
176645 |
0.04 |
| chr5_22853202_22853504 | 0.11 |
CDH12 |
cadherin 12, type 2 (N-cadherin 2) |
97 |
0.99 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.2 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.3 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.3 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.3 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.2 | GO:0036296 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.1 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0044803 | multi-organism membrane organization(GO:0044803) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0085029 | extracellular matrix assembly(GO:0085029) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0048488 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.0 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.0 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.0 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0033084 | immature T cell proliferation in thymus(GO:0033080) regulation of immature T cell proliferation in thymus(GO:0033084) negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.1 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |