Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
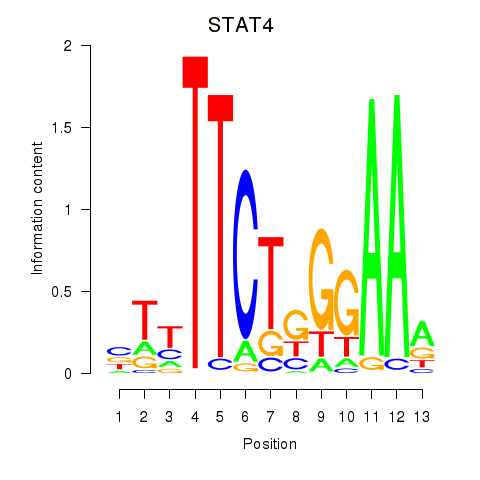
Results for STAT4
Z-value: 0.98
Transcription factors associated with STAT4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT4
|
ENSG00000138378.13 | signal transducer and activator of transcription 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_192029785_192029947 | STAT4 | 13544 | 0.219825 | 0.71 | 3.2e-02 | Click! |
| chr2_192037113_192037264 | STAT4 | 20866 | 0.205759 | 0.51 | 1.6e-01 | Click! |
| chr2_192058910_192059074 | STAT4 | 42670 | 0.151188 | -0.50 | 1.7e-01 | Click! |
| chr2_192014872_192015025 | STAT4 | 749 | 0.707304 | -0.49 | 1.8e-01 | Click! |
| chr2_192035909_192036060 | STAT4 | 19662 | 0.208262 | 0.43 | 2.5e-01 | Click! |
Activity of the STAT4 motif across conditions
Conditions sorted by the z-value of the STAT4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_190043861_190044108 | 0.57 |
COL5A2 |
collagen, type V, alpha 2 |
621 |
0.79 |
| chr5_54864628_54864779 | 0.57 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
33825 |
0.18 |
| chr8_32405226_32405421 | 0.51 |
NRG1 |
neuregulin 1 |
405 |
0.92 |
| chr10_33624842_33625153 | 0.48 |
NRP1 |
neuropilin 1 |
193 |
0.96 |
| chr20_55204347_55205219 | 0.42 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
425 |
0.81 |
| chr9_100069620_100070257 | 0.40 |
CCDC180 |
coiled-coil domain containing 180 |
3 |
0.98 |
| chr20_18720309_18720500 | 0.37 |
RP11-379J5.5 |
|
41051 |
0.15 |
| chr17_66951279_66951576 | 0.36 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
45 |
0.98 |
| chr7_19155354_19155672 | 0.34 |
TWIST1 |
twist family bHLH transcription factor 1 |
1782 |
0.28 |
| chr1_153584868_153585142 | 0.34 |
S100A16 |
S100 calcium binding protein A16 |
554 |
0.53 |
| chr4_81189966_81190509 | 0.34 |
FGF5 |
fibroblast growth factor 5 |
2444 |
0.36 |
| chr6_129203821_129204789 | 0.33 |
LAMA2 |
laminin, alpha 2 |
37 |
0.99 |
| chr4_126237285_126237744 | 0.33 |
FAT4 |
FAT atypical cadherin 4 |
40 |
0.99 |
| chr6_107816329_107816751 | 0.33 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
5378 |
0.23 |
| chr10_114820084_114820658 | 0.33 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
83308 |
0.1 |
| chr5_38846105_38846894 | 0.32 |
OSMR |
oncostatin M receptor |
398 |
0.91 |
| chr2_230134871_230135167 | 0.32 |
PID1 |
phosphotyrosine interaction domain containing 1 |
962 |
0.67 |
| chr14_52074790_52074946 | 0.31 |
ENSG00000251771 |
. |
3250 |
0.28 |
| chr20_10720180_10720390 | 0.30 |
JAG1 |
jagged 1 |
65591 |
0.13 |
| chr6_26745610_26745845 | 0.29 |
ZNF322 |
zinc finger protein 322 |
85747 |
0.08 |
| chr22_44726750_44727214 | 0.29 |
KIAA1644 |
KIAA1644 |
18251 |
0.23 |
| chr3_185960084_185960372 | 0.29 |
DGKG |
diacylglycerol kinase, gamma 90kDa |
38275 |
0.19 |
| chr2_38265076_38265999 | 0.28 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
2053 |
0.33 |
| chr1_182994808_182995119 | 0.28 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
2368 |
0.3 |
| chr13_30002091_30002575 | 0.28 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
431 |
0.89 |
| chr5_87436294_87437103 | 0.28 |
TMEM161B |
transmembrane protein 161B |
79750 |
0.11 |
| chr8_42358295_42358694 | 0.28 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
267 |
0.92 |
| chr12_117042205_117042356 | 0.27 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
28624 |
0.22 |
| chr9_97817794_97817986 | 0.27 |
ENSG00000207563 |
. |
29600 |
0.12 |
| chr6_74564945_74565311 | 0.27 |
RP11-553A21.3 |
|
159274 |
0.04 |
| chr2_42276471_42276770 | 0.27 |
PKDCC |
protein kinase domain containing, cytoplasmic |
1460 |
0.46 |
| chr7_152403601_152404275 | 0.26 |
XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
30688 |
0.14 |
| chr11_119953003_119953290 | 0.25 |
TRIM29 |
tripartite motif containing 29 |
38462 |
0.18 |
| chr3_180397078_180397946 | 0.25 |
CCDC39 |
coiled-coil domain containing 39 |
224 |
0.65 |
| chr11_122026624_122027083 | 0.25 |
ENSG00000207994 |
. |
3837 |
0.21 |
| chr8_495850_496251 | 0.25 |
TDRP |
testis development related protein |
269 |
0.95 |
| chr1_39572227_39572378 | 0.25 |
MACF1 |
microtubule-actin crosslinking factor 1 |
1219 |
0.45 |
| chr17_48546417_48546572 | 0.24 |
CHAD |
chondroadherin |
167 |
0.91 |
| chr2_190272487_190272871 | 0.24 |
WDR75 |
WD repeat domain 75 |
33480 |
0.21 |
| chr1_216897072_216897310 | 0.24 |
ESRRG |
estrogen-related receptor gamma |
396 |
0.93 |
| chr3_64199785_64199936 | 0.24 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
11271 |
0.21 |
| chr8_134516643_134516794 | 0.24 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
5092 |
0.34 |
| chr11_8854095_8854571 | 0.24 |
ST5 |
suppression of tumorigenicity 5 |
847 |
0.53 |
| chr4_187491209_187491867 | 0.24 |
MTNR1A |
melatonin receptor 1A |
14817 |
0.17 |
| chr9_80981008_80981159 | 0.23 |
PSAT1 |
phosphoserine aminotransferase 1 |
69024 |
0.13 |
| chr18_31336109_31336260 | 0.23 |
ASXL3 |
additional sex combs like 3 (Drosophila) |
150634 |
0.04 |
| chr2_138721657_138722065 | 0.23 |
HNMT |
histamine N-methyltransferase |
19 |
0.99 |
| chr15_38693567_38693718 | 0.23 |
FAM98B |
family with sequence similarity 98, member B |
52686 |
0.15 |
| chr4_128703513_128704516 | 0.23 |
HSPA4L |
heat shock 70kDa protein 4-like |
537 |
0.81 |
| chr10_101548471_101548669 | 0.23 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
6050 |
0.22 |
| chr11_12029509_12029738 | 0.22 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
507 |
0.87 |
| chr22_46546744_46547896 | 0.22 |
PPARA |
peroxisome proliferator-activated receptor alpha |
467 |
0.75 |
| chr11_46237812_46237970 | 0.22 |
CTD-2589M5.4 |
|
58207 |
0.1 |
| chr5_64558662_64558851 | 0.22 |
ENSG00000207439 |
. |
139560 |
0.05 |
| chr1_109401493_109401651 | 0.22 |
SPATA42 |
spermatogenesis associated 42 (non-protein coding) |
1706 |
0.28 |
| chr20_12989594_12990369 | 0.22 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
16 |
0.99 |
| chr10_17276996_17277147 | 0.22 |
RP11-124N14.3 |
|
239 |
0.91 |
| chr7_155006728_155006879 | 0.22 |
AC099552.4 |
Uncharacterized protein |
16657 |
0.2 |
| chr15_76004107_76004336 | 0.22 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
968 |
0.37 |
| chr10_116444307_116444502 | 0.22 |
ABLIM1 |
actin binding LIM protein 1 |
10 |
0.98 |
| chr4_157872609_157873145 | 0.22 |
PDGFC |
platelet derived growth factor C |
19178 |
0.21 |
| chr11_109816668_109816859 | 0.22 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
147324 |
0.05 |
| chr17_53799956_53800183 | 0.22 |
TMEM100 |
transmembrane protein 100 |
148 |
0.97 |
| chr8_17554662_17554991 | 0.22 |
MTUS1 |
microtubule associated tumor suppressor 1 |
353 |
0.86 |
| chr6_26780154_26780414 | 0.22 |
GUSBP2 |
glucuronidase, beta pseudogene 2 |
66333 |
0.11 |
| chr2_208378036_208378402 | 0.21 |
CREB1 |
cAMP responsive element binding protein 1 |
16242 |
0.2 |
| chr17_1665908_1666244 | 0.21 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
47 |
0.96 |
| chr1_48935703_48936400 | 0.21 |
SPATA6 |
spermatogenesis associated 6 |
1631 |
0.46 |
| chr2_14775112_14775752 | 0.21 |
AC011897.1 |
Uncharacterized protein |
217 |
0.95 |
| chr3_99359780_99359931 | 0.21 |
COL8A1 |
collagen, type VIII, alpha 1 |
2401 |
0.39 |
| chr19_42617987_42618295 | 0.21 |
POU2F2 |
POU class 2 homeobox 2 |
2898 |
0.14 |
| chr8_122335916_122336137 | 0.21 |
ENSG00000221644 |
. |
136894 |
0.05 |
| chr3_157156592_157156743 | 0.21 |
PTX3 |
pentraxin 3, long |
2089 |
0.41 |
| chr21_44739435_44739605 | 0.21 |
SIK1 |
salt-inducible kinase 1 |
107488 |
0.06 |
| chr6_125285262_125285420 | 0.21 |
RP11-510H23.1 |
|
1156 |
0.45 |
| chr2_67801207_67801358 | 0.21 |
ETAA1 |
Ewing tumor-associated antigen 1 |
176831 |
0.03 |
| chrX_114467670_114468385 | 0.21 |
LRCH2 |
leucine-rich repeats and calponin homology (CH) domain containing 2 |
601 |
0.77 |
| chr5_110429004_110429186 | 0.21 |
WDR36 |
WD repeat domain 36 |
1076 |
0.41 |
| chr10_131458318_131458596 | 0.20 |
RP11-109A6.2 |
|
41069 |
0.2 |
| chr3_116162193_116162478 | 0.20 |
LSAMP |
limbic system-associated membrane protein |
1495 |
0.53 |
| chr11_76495074_76495334 | 0.20 |
TSKU |
tsukushi, small leucine rich proteoglycan |
12 |
0.95 |
| chr10_104040372_104040852 | 0.20 |
ENSG00000251989 |
. |
19625 |
0.14 |
| chr11_126289852_126290003 | 0.20 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
13250 |
0.18 |
| chr5_158488269_158488591 | 0.20 |
EBF1 |
early B-cell factor 1 |
38271 |
0.17 |
| chr5_50682748_50683080 | 0.20 |
ISL1 |
ISL LIM homeobox 1 |
3408 |
0.27 |
| chr11_7507047_7507406 | 0.20 |
OLFML1 |
olfactomedin-like 1 |
389 |
0.86 |
| chr5_111017573_111017756 | 0.20 |
ENSG00000253057 |
. |
38094 |
0.17 |
| chr2_189842947_189843104 | 0.20 |
ENSG00000221502 |
. |
207 |
0.95 |
| chr18_67067194_67067650 | 0.20 |
DOK6 |
docking protein 6 |
869 |
0.76 |
| chr12_104360763_104360914 | 0.20 |
TDG |
thymine-DNA glycosylase |
1174 |
0.31 |
| chr2_48739670_48739821 | 0.20 |
ENSG00000202227 |
. |
10582 |
0.18 |
| chr8_128982063_128982325 | 0.20 |
ENSG00000221771 |
. |
9315 |
0.18 |
| chr3_99356620_99357126 | 0.19 |
COL8A1 |
collagen, type VIII, alpha 1 |
446 |
0.89 |
| chr11_122070808_122070959 | 0.19 |
ENSG00000207994 |
. |
47867 |
0.13 |
| chr4_75859213_75859706 | 0.19 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
1133 |
0.59 |
| chr6_143501955_143502106 | 0.19 |
AIG1 |
androgen-induced 1 |
54643 |
0.14 |
| chr8_68896954_68897148 | 0.19 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
32698 |
0.23 |
| chr12_110720685_110720925 | 0.19 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
1319 |
0.51 |
| chr2_235541415_235541566 | 0.19 |
ARL4C |
ADP-ribosylation factor-like 4C |
135793 |
0.05 |
| chr10_27976466_27976617 | 0.19 |
MKX |
mohawk homeobox |
55933 |
0.13 |
| chr8_62602537_62602767 | 0.19 |
ASPH |
aspartate beta-hydroxylase |
244 |
0.94 |
| chr13_47070209_47070436 | 0.19 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
56981 |
0.12 |
| chr21_30178047_30178198 | 0.19 |
ENSG00000251894 |
. |
62592 |
0.12 |
| chr2_176931780_176932252 | 0.19 |
EVX2 |
even-skipped homeobox 2 |
16625 |
0.1 |
| chr7_116312981_116313724 | 0.19 |
MET |
met proto-oncogene |
893 |
0.68 |
| chr2_175711229_175711408 | 0.19 |
CHN1 |
chimerin 1 |
175 |
0.97 |
| chr14_33750234_33750385 | 0.18 |
NPAS3 |
neuronal PAS domain protein 3 |
65791 |
0.15 |
| chr4_24472317_24472597 | 0.18 |
ENSG00000265001 |
. |
41002 |
0.15 |
| chr1_201600215_201600366 | 0.18 |
NAV1 |
neuron navigator 1 |
7689 |
0.16 |
| chr3_129407054_129407730 | 0.18 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
183 |
0.96 |
| chr1_227413559_227414076 | 0.18 |
RP11-1B20.1 |
|
47783 |
0.17 |
| chr2_178938029_178938202 | 0.18 |
PDE11A |
phosphodiesterase 11A |
633 |
0.72 |
| chr8_127837007_127837644 | 0.18 |
ENSG00000212451 |
. |
153558 |
0.04 |
| chr10_16535131_16535282 | 0.18 |
PTER |
phosphotriesterase related |
8875 |
0.2 |
| chr4_177714164_177714554 | 0.18 |
VEGFC |
vascular endothelial growth factor C |
478 |
0.89 |
| chr7_41735476_41735627 | 0.18 |
INHBA-AS1 |
INHBA antisense RNA 1 |
2005 |
0.33 |
| chr4_138449879_138450179 | 0.18 |
PCDH18 |
protocadherin 18 |
3536 |
0.4 |
| chr6_132691040_132691268 | 0.18 |
MOXD1 |
monooxygenase, DBH-like 1 |
5050 |
0.31 |
| chr6_56707311_56707814 | 0.18 |
DST |
dystonin |
381 |
0.81 |
| chr1_157980267_157980690 | 0.18 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
14862 |
0.2 |
| chr5_130300100_130300251 | 0.18 |
HINT1 |
histidine triad nucleotide binding protein 1 |
198145 |
0.03 |
| chr8_134276506_134276691 | 0.18 |
NDRG1 |
N-myc downstream regulated 1 |
32181 |
0.18 |
| chr3_178816583_178816734 | 0.18 |
ENSG00000200616 |
. |
1139 |
0.51 |
| chr11_12701583_12701887 | 0.18 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
4685 |
0.33 |
| chr5_17365786_17365937 | 0.18 |
ENSG00000201715 |
. |
20136 |
0.26 |
| chr14_63670391_63670709 | 0.18 |
RHOJ |
ras homolog family member J |
576 |
0.81 |
| chr4_162286727_162286878 | 0.18 |
RP11-234O6.2 |
|
13303 |
0.32 |
| chr8_18869881_18870079 | 0.18 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
1216 |
0.52 |
| chr10_63551124_63551275 | 0.17 |
RP11-63A2.2 |
|
106053 |
0.07 |
| chr1_14220028_14220179 | 0.17 |
PRDM2 |
PR domain containing 2, with ZNF domain |
121882 |
0.06 |
| chr11_65662993_65663440 | 0.17 |
FOSL1 |
FOS-like antigen 1 |
4674 |
0.08 |
| chr3_149271657_149271885 | 0.17 |
WWTR1 |
WW domain containing transcription regulator 1 |
22268 |
0.17 |
| chr7_54972295_54972520 | 0.17 |
ENSG00000252054 |
. |
38896 |
0.2 |
| chr6_131019035_131019186 | 0.17 |
ENSG00000202438 |
. |
123853 |
0.06 |
| chr9_4144735_4144971 | 0.17 |
GLIS3 |
GLIS family zinc finger 3 |
340 |
0.93 |
| chr6_163840513_163841070 | 0.17 |
QKI |
QKI, KH domain containing, RNA binding |
3375 |
0.39 |
| chr5_43075817_43076018 | 0.17 |
ZNF131 |
zinc finger protein 131 |
7758 |
0.16 |
| chr8_118056802_118056966 | 0.17 |
ENSG00000244033 |
. |
26699 |
0.2 |
| chr2_235403849_235404784 | 0.17 |
ARL4C |
ADP-ribosylation factor-like 4C |
928 |
0.74 |
| chr2_178126484_178126695 | 0.17 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
1593 |
0.23 |
| chr15_60682576_60682790 | 0.17 |
ANXA2 |
annexin A2 |
643 |
0.8 |
| chr12_22005080_22005298 | 0.17 |
RP11-729I10.2 |
|
25045 |
0.19 |
| chr6_138427102_138427253 | 0.17 |
PERP |
PERP, TP53 apoptosis effector |
1471 |
0.51 |
| chr3_175244326_175245006 | 0.17 |
ENSG00000201648 |
. |
82601 |
0.1 |
| chr18_8140189_8140340 | 0.17 |
RP11-376P6.3 |
|
14292 |
0.21 |
| chr4_88419376_88419544 | 0.17 |
SPARCL1 |
SPARC-like 1 (hevin) |
30311 |
0.17 |
| chr3_179244742_179244893 | 0.17 |
ACTL6A |
actin-like 6A |
35851 |
0.12 |
| chr2_222366683_222366911 | 0.17 |
EPHA4 |
EPH receptor A4 |
483 |
0.85 |
| chr3_73776246_73776397 | 0.17 |
PDZRN3 |
PDZ domain containing ring finger 3 |
102230 |
0.07 |
| chr4_54644503_54644654 | 0.17 |
LNX1 |
ligand of numb-protein X 1, E3 ubiquitin protein ligase |
77006 |
0.1 |
| chr6_82767477_82767628 | 0.17 |
ENSG00000223044 |
. |
152605 |
0.04 |
| chr9_684288_685148 | 0.17 |
RP11-130C19.3 |
|
837 |
0.67 |
| chr11_133990232_133990720 | 0.17 |
NCAPD3 |
non-SMC condensin II complex, subunit D3 |
47891 |
0.12 |
| chr1_183841636_183841838 | 0.17 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
67447 |
0.11 |
| chr6_28952769_28953226 | 0.17 |
HCG15 |
HLA complex group 15 (non-protein coding) |
983 |
0.36 |
| chr2_189842795_189842946 | 0.17 |
ENSG00000221502 |
. |
52 |
0.98 |
| chrX_19701596_19701747 | 0.17 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
12555 |
0.29 |
| chr8_136471145_136471318 | 0.17 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
281 |
0.92 |
| chr2_6963326_6963477 | 0.16 |
AC017076.5 |
|
10261 |
0.21 |
| chr2_197345735_197345886 | 0.16 |
HECW2 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
111525 |
0.06 |
| chr5_159292030_159292306 | 0.16 |
ADRA1B |
adrenoceptor alpha 1B |
51622 |
0.16 |
| chr12_26303991_26304142 | 0.16 |
SSPN |
sarcospan |
16166 |
0.18 |
| chr1_196966907_196967058 | 0.16 |
CFHR5 |
complement factor H-related 5 |
20296 |
0.2 |
| chr14_69310573_69310778 | 0.16 |
ENSG00000263694 |
. |
1844 |
0.38 |
| chr2_145266987_145267316 | 0.16 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
7964 |
0.25 |
| chr8_124400004_124400155 | 0.16 |
ATAD2 |
ATPase family, AAA domain containing 2 |
8626 |
0.16 |
| chr11_89113858_89114276 | 0.16 |
NOX4 |
NADPH oxidase 4 |
109833 |
0.07 |
| chr22_44982741_44982930 | 0.16 |
PRR5 |
proline rich 5 (renal) |
81758 |
0.09 |
| chr16_75298860_75299117 | 0.16 |
BCAR1 |
breast cancer anti-estrogen resistance 1 |
125 |
0.95 |
| chr9_133656541_133656767 | 0.16 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
53799 |
0.13 |
| chr1_108115540_108115852 | 0.16 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
115430 |
0.06 |
| chr1_156644232_156644383 | 0.16 |
NES |
nestin |
2882 |
0.13 |
| chr17_77767116_77767731 | 0.16 |
CBX8 |
chromobox homolog 8 |
3492 |
0.16 |
| chr1_1207989_1209115 | 0.16 |
UBE2J2 |
ubiquitin-conjugating enzyme E2, J2 |
299 |
0.76 |
| chr4_99575467_99575704 | 0.16 |
TSPAN5 |
tetraspanin 5 |
3201 |
0.28 |
| chr1_60204181_60204332 | 0.16 |
ENSG00000266150 |
. |
5288 |
0.28 |
| chr3_187121306_187121457 | 0.16 |
ENSG00000239093 |
. |
19722 |
0.21 |
| chr16_51360009_51360198 | 0.16 |
SALL1 |
spalt-like transcription factor 1 |
174917 |
0.03 |
| chr1_32930228_32931040 | 0.16 |
ZBTB8B |
zinc finger and BTB domain containing 8B |
36 |
0.97 |
| chr6_116718983_116719178 | 0.16 |
DSE |
dermatan sulfate epimerase |
26970 |
0.15 |
| chr1_120191467_120191618 | 0.16 |
ZNF697 |
zinc finger protein 697 |
1146 |
0.52 |
| chr5_14157312_14157506 | 0.16 |
TRIO |
trio Rho guanine nucleotide exchange factor |
13580 |
0.31 |
| chr9_97906510_97906877 | 0.16 |
ENSG00000251884 |
. |
1870 |
0.3 |
| chr2_175470788_175470939 | 0.16 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
7924 |
0.2 |
| chr6_53875762_53875944 | 0.16 |
MLIP |
muscular LMNA-interacting protein |
7861 |
0.2 |
| chr17_32078348_32078506 | 0.16 |
ENSG00000221699 |
. |
123050 |
0.05 |
| chr17_1666268_1666531 | 0.16 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
276 |
0.85 |
| chr9_110500431_110500703 | 0.16 |
AL162389.1 |
Uncharacterized protein |
39852 |
0.16 |
| chr2_20133345_20133656 | 0.16 |
WDR35 |
WD repeat domain 35 |
20132 |
0.18 |
| chr1_39658092_39658302 | 0.16 |
MACF1 |
microtubule-actin crosslinking factor 1 |
11721 |
0.17 |
| chr17_46672091_46672242 | 0.16 |
HOXB5 |
homeobox B5 |
843 |
0.28 |
| chr13_20875473_20875869 | 0.15 |
GJB6 |
gap junction protein, beta 6, 30kDa |
69137 |
0.11 |
| chr8_59746213_59746613 | 0.15 |
ENSG00000201231 |
. |
19302 |
0.27 |
| chr8_116588884_116589247 | 0.15 |
TRPS1 |
trichorhinophalangeal syndrome I |
43232 |
0.19 |
| chr14_92413096_92413399 | 0.15 |
FBLN5 |
fibulin 5 |
171 |
0.96 |
| chr18_5895024_5895391 | 0.15 |
TMEM200C |
transmembrane protein 200C |
747 |
0.66 |
| chr6_83076044_83076195 | 0.15 |
TPBG |
trophoblast glycoprotein |
2158 |
0.46 |
| chr7_82070146_82070432 | 0.15 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
2742 |
0.43 |
| chr12_131438586_131438737 | 0.15 |
GPR133 |
G protein-coupled receptor 133 |
137 |
0.97 |
| chr8_93113958_93114847 | 0.15 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
1052 |
0.69 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.5 | GO:0010658 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.3 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.3 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.2 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.1 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.2 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.1 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.2 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.2 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.0 | GO:0009415 | response to water deprivation(GO:0009414) response to water(GO:0009415) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.0 | 0.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.2 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.1 | GO:0060008 | Sertoli cell differentiation(GO:0060008) Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.4 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.2 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:1990748 | removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0045008 | depyrimidination(GO:0045008) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0072193 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0071385 | cellular response to corticosteroid stimulus(GO:0071384) cellular response to glucocorticoid stimulus(GO:0071385) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.3 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.1 | 0.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.1 | 0.2 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.1 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.3 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.2 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.0 | SIG CHEMOTAXIS | Genes related to chemotaxis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |