Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
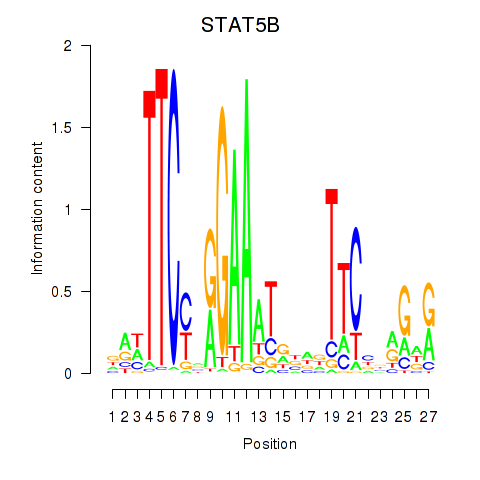
Results for STAT5B
Z-value: 0.61
Transcription factors associated with STAT5B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT5B
|
ENSG00000173757.5 | signal transducer and activator of transcription 5B |
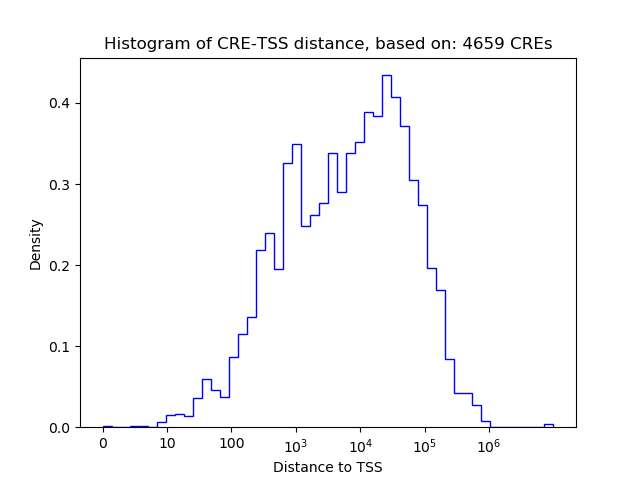
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_40429185_40429364 | STAT5B | 549 | 0.641731 | -0.43 | 2.5e-01 | Click! |
| chr17_40426926_40427118 | STAT5B | 1402 | 0.266467 | -0.43 | 2.5e-01 | Click! |
| chr17_40427243_40427997 | STAT5B | 804 | 0.471429 | -0.31 | 4.2e-01 | Click! |
| chr17_40428589_40429042 | STAT5B | 90 | 0.946452 | -0.27 | 4.9e-01 | Click! |
| chr17_40428204_40428355 | STAT5B | 145 | 0.924914 | -0.11 | 7.7e-01 | Click! |
Activity of the STAT5B motif across conditions
Conditions sorted by the z-value of the STAT5B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
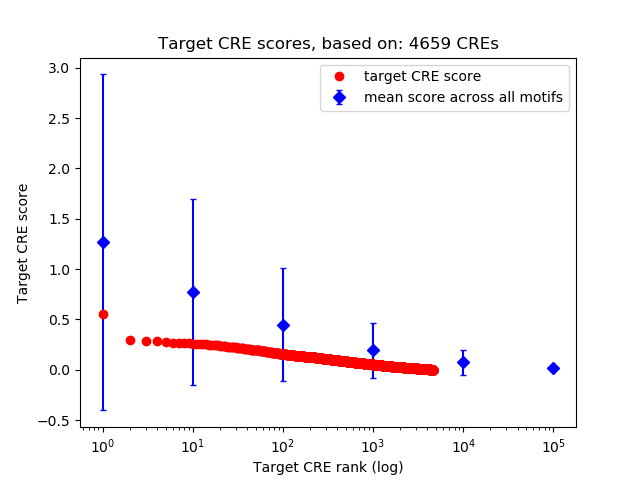
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_138449879_138450179 | 0.55 |
PCDH18 |
protocadherin 18 |
3536 |
0.4 |
| chr19_44860491_44860902 | 0.29 |
ZNF112 |
zinc finger protein 112 |
130 |
0.96 |
| chr12_56138916_56139328 | 0.29 |
GDF11 |
growth differentiation factor 11 |
1939 |
0.16 |
| chr10_126843486_126843769 | 0.28 |
CTBP2 |
C-terminal binding protein 2 |
3658 |
0.35 |
| chr17_59478534_59478762 | 0.27 |
RP11-332H18.5 |
|
1044 |
0.31 |
| chr9_115774055_115774548 | 0.27 |
ZFP37 |
ZFP37 zinc finger protein |
44667 |
0.16 |
| chr7_112725821_112726406 | 0.27 |
GPR85 |
G protein-coupled receptor 85 |
280 |
0.92 |
| chr19_55684291_55685412 | 0.27 |
SYT5 |
synaptotagmin V |
1083 |
0.28 |
| chr6_27515697_27516098 | 0.26 |
ENSG00000206671 |
. |
48294 |
0.13 |
| chr5_38846105_38846894 | 0.26 |
OSMR |
oncostatin M receptor |
398 |
0.91 |
| chr19_55677599_55678034 | 0.26 |
DNAAF3 |
dynein, axonemal, assembly factor 3 |
104 |
0.87 |
| chr1_172110530_172111247 | 0.26 |
ENSG00000207949 |
. |
2841 |
0.22 |
| chr2_133061942_133062584 | 0.26 |
ENSG00000221288 |
. |
47610 |
0.14 |
| chr10_22894404_22894761 | 0.26 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
13940 |
0.29 |
| chr3_129407054_129407730 | 0.25 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
183 |
0.96 |
| chr15_63337269_63337696 | 0.25 |
TPM1 |
tropomyosin 1 (alpha) |
1507 |
0.37 |
| chr8_105373378_105373573 | 0.25 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
12706 |
0.2 |
| chr13_27557206_27557734 | 0.24 |
USP12-AS1 |
USP12 antisense RNA 1 |
179522 |
0.03 |
| chr3_123602332_123603013 | 0.24 |
MYLK |
myosin light chain kinase |
477 |
0.85 |
| chr19_46916092_46916684 | 0.24 |
CCDC8 |
coiled-coil domain containing 8 |
453 |
0.75 |
| chr17_12568379_12568897 | 0.23 |
MYOCD |
myocardin |
668 |
0.75 |
| chr4_160219678_160219941 | 0.23 |
ENSG00000216089 |
. |
2943 |
0.31 |
| chr15_44093610_44093878 | 0.23 |
HYPK |
huntingtin interacting protein K |
947 |
0.25 |
| chrX_67913478_67914353 | 0.23 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
422 |
0.91 |
| chr7_152403601_152404275 | 0.23 |
XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
30688 |
0.14 |
| chr11_8854095_8854571 | 0.23 |
ST5 |
suppression of tumorigenicity 5 |
847 |
0.53 |
| chr11_57530814_57530965 | 0.23 |
CTNND1 |
catenin (cadherin-associated protein), delta 1 |
436 |
0.77 |
| chr9_97662711_97663102 | 0.22 |
RP11-49O14.2 |
|
195 |
0.95 |
| chr2_223163063_223163341 | 0.22 |
PAX3 |
paired box 3 |
263 |
0.57 |
| chr10_89802413_89802899 | 0.22 |
ENSG00000200891 |
. |
48204 |
0.17 |
| chr7_47576453_47576928 | 0.22 |
TNS3 |
tensin 3 |
2185 |
0.45 |
| chr8_127846488_127846716 | 0.22 |
ENSG00000212451 |
. |
162835 |
0.04 |
| chr1_95330057_95330663 | 0.22 |
SLC44A3 |
solute carrier family 44, member 3 |
2527 |
0.28 |
| chr4_118005872_118006140 | 0.22 |
TRAM1L1 |
translocation associated membrane protein 1-like 1 |
730 |
0.82 |
| chr8_68304896_68305241 | 0.22 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
49156 |
0.18 |
| chr7_132299406_132299928 | 0.21 |
PLXNA4 |
plexin A4 |
33780 |
0.18 |
| chr6_163840513_163841070 | 0.21 |
QKI |
QKI, KH domain containing, RNA binding |
3375 |
0.39 |
| chr9_19493377_19493724 | 0.21 |
RP11-363E7.4 |
|
40343 |
0.16 |
| chr2_151336714_151337073 | 0.21 |
RND3 |
Rho family GTPase 3 |
5003 |
0.36 |
| chr4_54063057_54063462 | 0.21 |
ENSG00000207385 |
. |
68475 |
0.13 |
| chr10_36461738_36461970 | 0.21 |
ENSG00000237002 |
. |
265735 |
0.02 |
| chr3_45096456_45096713 | 0.20 |
ENSG00000252410 |
. |
20504 |
0.16 |
| chr7_100765722_100766153 | 0.20 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
4433 |
0.12 |
| chr15_33009221_33009412 | 0.20 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
859 |
0.63 |
| chr13_114147905_114148103 | 0.20 |
TMCO3 |
transmembrane and coiled-coil domains 3 |
2646 |
0.19 |
| chr7_143581670_143581927 | 0.20 |
FAM115A |
family with sequence similarity 115, member A |
665 |
0.7 |
| chr5_7396416_7397106 | 0.20 |
ADCY2 |
adenylate cyclase 2 (brain) |
440 |
0.88 |
| chr2_159992813_159993143 | 0.20 |
ENSG00000202029 |
. |
109324 |
0.06 |
| chr6_129203821_129204789 | 0.20 |
LAMA2 |
laminin, alpha 2 |
37 |
0.99 |
| chr3_167812396_167812591 | 0.20 |
GOLIM4 |
golgi integral membrane protein 4 |
639 |
0.83 |
| chr12_10902635_10902853 | 0.20 |
YBX3 |
Y box binding protein 3 |
26833 |
0.13 |
| chr15_37175433_37176047 | 0.20 |
ENSG00000212511 |
. |
30899 |
0.23 |
| chr13_73438368_73438629 | 0.20 |
ENSG00000220981 |
. |
13272 |
0.19 |
| chr3_61649650_61649813 | 0.19 |
ENSG00000252420 |
. |
75172 |
0.12 |
| chr5_82768204_82768925 | 0.19 |
VCAN |
versican |
820 |
0.75 |
| chr12_15941079_15941239 | 0.19 |
EPS8 |
epidermal growth factor receptor pathway substrate 8 |
1156 |
0.62 |
| chr1_54197803_54198068 | 0.19 |
GLIS1 |
GLIS family zinc finger 1 |
1942 |
0.34 |
| chr9_21764802_21765039 | 0.19 |
MTAP |
methylthioadenosine phosphorylase |
37622 |
0.15 |
| chr7_155006728_155006879 | 0.19 |
AC099552.4 |
Uncharacterized protein |
16657 |
0.2 |
| chr11_79149678_79149969 | 0.19 |
TENM4 |
teneurin transmembrane protein 4 |
1872 |
0.38 |
| chr3_149094469_149095370 | 0.19 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
646 |
0.47 |
| chr2_142888867_142889088 | 0.18 |
LRP1B |
low density lipoprotein receptor-related protein 1B |
293 |
0.93 |
| chr11_123062062_123062213 | 0.18 |
CTD-2216M2.1 |
|
2627 |
0.27 |
| chr2_145266428_145266579 | 0.18 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
8612 |
0.25 |
| chr20_23224910_23225093 | 0.18 |
ENSG00000201527 |
. |
83393 |
0.07 |
| chr15_42749459_42749764 | 0.18 |
ZNF106 |
zinc finger protein 106 |
100 |
0.96 |
| chr10_17034557_17035249 | 0.18 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
8709 |
0.3 |
| chr7_77650153_77650326 | 0.18 |
RP5-1185I7.1 |
|
30502 |
0.24 |
| chr1_221509112_221509492 | 0.17 |
DUSP10 |
dual specificity phosphatase 10 |
401500 |
0.01 |
| chr8_136471145_136471318 | 0.17 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
281 |
0.92 |
| chr1_77737435_77737712 | 0.17 |
AK5 |
adenylate kinase 5 |
10163 |
0.23 |
| chr11_89113858_89114276 | 0.17 |
NOX4 |
NADPH oxidase 4 |
109833 |
0.07 |
| chr12_29935925_29936182 | 0.17 |
TMTC1 |
transmembrane and tetratricopeptide repeat containing 1 |
678 |
0.82 |
| chr11_122311553_122311819 | 0.17 |
ENSG00000252776 |
. |
8232 |
0.23 |
| chr3_137833372_137833843 | 0.17 |
DZIP1L |
DAZ interacting zinc finger protein 1-like |
844 |
0.63 |
| chr20_14317720_14318261 | 0.17 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
264 |
0.94 |
| chr5_43308499_43308850 | 0.17 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
4652 |
0.23 |
| chr2_190477763_190477914 | 0.17 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
29354 |
0.16 |
| chrX_106045541_106045714 | 0.17 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
283 |
0.94 |
| chr1_87793556_87794116 | 0.17 |
LMO4 |
LIM domain only 4 |
315 |
0.94 |
| chr17_46672091_46672242 | 0.17 |
HOXB5 |
homeobox B5 |
843 |
0.28 |
| chr6_113700515_113700666 | 0.17 |
ENSG00000222677 |
. |
135599 |
0.05 |
| chr20_17643261_17643428 | 0.17 |
RRBP1 |
ribosome binding protein 1 |
2192 |
0.33 |
| chr5_89317325_89317513 | 0.17 |
ENSG00000264342 |
. |
4882 |
0.36 |
| chr11_114167823_114168031 | 0.17 |
NNMT |
nicotinamide N-methyltransferase |
158 |
0.96 |
| chr1_196620715_196621368 | 0.17 |
CFH |
complement factor H |
33 |
0.98 |
| chr5_123493061_123493232 | 0.16 |
CSNK1G3 |
casein kinase 1, gamma 3 |
569027 |
0.0 |
| chr8_104384541_104384884 | 0.16 |
CTHRC1 |
collagen triple helix repeat containing 1 |
51 |
0.97 |
| chr3_161883288_161883607 | 0.16 |
OTOL1 |
otolin 1 |
668851 |
0.0 |
| chr6_27205685_27206711 | 0.16 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chr3_33397150_33397316 | 0.16 |
ENSG00000252700 |
. |
58409 |
0.12 |
| chr2_158752022_158752173 | 0.16 |
ENSG00000251980 |
. |
17187 |
0.16 |
| chr5_83140533_83140765 | 0.16 |
ENSG00000202157 |
. |
41276 |
0.19 |
| chr18_60052258_60052436 | 0.16 |
RP11-640A1.3 |
|
6984 |
0.22 |
| chr16_18811323_18811474 | 0.16 |
ARL6IP1 |
ADP-ribosylation factor-like 6 interacting protein 1 |
1348 |
0.27 |
| chr19_58400508_58400670 | 0.16 |
ZNF814 |
zinc finger protein 814 |
184 |
0.9 |
| chr2_151340168_151340387 | 0.16 |
RND3 |
Rho family GTPase 3 |
1619 |
0.57 |
| chr5_122620941_122621184 | 0.16 |
CEP120 |
centrosomal protein 120kDa |
137935 |
0.04 |
| chr7_38624277_38624485 | 0.16 |
AMPH |
amphiphysin |
46639 |
0.16 |
| chr5_119874213_119874384 | 0.15 |
PRR16 |
proline rich 16 |
7139 |
0.33 |
| chr8_17106304_17106459 | 0.15 |
VPS37A |
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
1628 |
0.35 |
| chr6_16675168_16675421 | 0.15 |
RP1-151F17.1 |
|
86075 |
0.1 |
| chr6_27173563_27173801 | 0.15 |
PRSS16 |
protease, serine, 16 (thymus) |
41798 |
0.1 |
| chr8_94238484_94238635 | 0.15 |
AC016885.1 |
Uncharacterized protein |
3308 |
0.29 |
| chr19_39919352_39919568 | 0.15 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
4049 |
0.09 |
| chr2_145212550_145212714 | 0.15 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
24495 |
0.24 |
| chr2_37116896_37117561 | 0.15 |
ENSG00000252756 |
. |
22590 |
0.18 |
| chr1_231543825_231544251 | 0.15 |
EGLN1 |
egl-9 family hypoxia-inducible factor 1 |
16752 |
0.2 |
| chr2_232492715_232492900 | 0.15 |
ENSG00000239202 |
. |
18177 |
0.16 |
| chr8_54793517_54794385 | 0.15 |
RGS20 |
regulator of G-protein signaling 20 |
497 |
0.79 |
| chr10_5491792_5492244 | 0.15 |
NET1 |
neuroepithelial cell transforming 1 |
3444 |
0.22 |
| chr1_236827439_236827590 | 0.15 |
ACTN2 |
actinin, alpha 2 |
22240 |
0.21 |
| chr1_201916514_201916665 | 0.15 |
LMOD1 |
leiomodin 1 (smooth muscle) |
874 |
0.44 |
| chr13_80707939_80708090 | 0.15 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
205780 |
0.03 |
| chr6_27584810_27585278 | 0.15 |
ENSG00000238648 |
. |
14144 |
0.19 |
| chr8_77867156_77867509 | 0.15 |
ENSG00000266712 |
. |
11754 |
0.26 |
| chr15_96886136_96886425 | 0.15 |
ENSG00000222651 |
. |
9790 |
0.16 |
| chrX_28604967_28605213 | 0.15 |
IL1RAPL1 |
interleukin 1 receptor accessory protein-like 1 |
426 |
0.92 |
| chr1_32268041_32268276 | 0.15 |
SPOCD1 |
SPOC domain containing 1 |
3718 |
0.19 |
| chr16_74204020_74204218 | 0.15 |
PSMD7 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
126554 |
0.05 |
| chr3_197099341_197099661 | 0.15 |
ENSG00000238491 |
. |
33079 |
0.17 |
| chr17_739288_739623 | 0.15 |
NXN |
nucleoredoxin |
10214 |
0.15 |
| chr8_125827427_125827626 | 0.15 |
ENSG00000263735 |
. |
6774 |
0.28 |
| chr20_49307514_49308405 | 0.15 |
FAM65C |
family with sequence similarity 65, member C |
43 |
0.97 |
| chr6_139656188_139656543 | 0.15 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
38985 |
0.15 |
| chr10_19026614_19026765 | 0.15 |
ARL5B |
ADP-ribosylation factor-like 5B |
78355 |
0.1 |
| chr6_27255999_27256495 | 0.15 |
POM121L2 |
POM121 transmembrane nucleoporin-like 2 |
22844 |
0.18 |
| chr8_108342593_108342744 | 0.15 |
ANGPT1 |
angiopoietin 1 |
6082 |
0.32 |
| chr12_105065797_105066143 | 0.15 |
ENSG00000264295 |
. |
80559 |
0.1 |
| chr6_116832868_116833176 | 0.15 |
FAM26E |
family with sequence similarity 26, member E |
213 |
0.73 |
| chr3_75690482_75691208 | 0.14 |
ENSG00000221795 |
. |
10931 |
0.2 |
| chr2_60808641_60809007 | 0.14 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
28122 |
0.19 |
| chr11_122513572_122513810 | 0.14 |
UBASH3B |
ubiquitin associated and SH3 domain containing B |
12692 |
0.24 |
| chr9_123698545_123698822 | 0.14 |
TRAF1 |
TNF receptor-associated factor 1 |
7232 |
0.23 |
| chr15_39871853_39872515 | 0.14 |
THBS1 |
thrombospondin 1 |
1096 |
0.52 |
| chrX_99904753_99905093 | 0.14 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
5708 |
0.19 |
| chr5_167696137_167696498 | 0.14 |
WWC1 |
WW and C2 domain containing 1 |
22339 |
0.2 |
| chr1_8587025_8587260 | 0.14 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
1056 |
0.58 |
| chr10_13748985_13749251 | 0.14 |
ENSG00000222235 |
. |
2140 |
0.21 |
| chr2_47499719_47500092 | 0.14 |
EPCAM |
epithelial cell adhesion molecule |
72392 |
0.09 |
| chr4_142003365_142003602 | 0.14 |
RNF150 |
ring finger protein 150 |
28603 |
0.25 |
| chr9_75653628_75653779 | 0.14 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
65 |
0.99 |
| chr15_98504600_98505000 | 0.14 |
ARRDC4 |
arrestin domain containing 4 |
872 |
0.68 |
| chr9_110250307_110251795 | 0.14 |
KLF4 |
Kruppel-like factor 4 (gut) |
360 |
0.89 |
| chr20_31063509_31063841 | 0.14 |
C20orf112 |
chromosome 20 open reading frame 112 |
7599 |
0.19 |
| chr5_54864628_54864779 | 0.14 |
PPAP2A |
phosphatidic acid phosphatase type 2A |
33825 |
0.18 |
| chr10_14032873_14033043 | 0.14 |
RP11-142M10.2 |
|
857 |
0.66 |
| chr3_55519561_55519953 | 0.14 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1574 |
0.4 |
| chr19_44598113_44598899 | 0.14 |
ZNF224 |
zinc finger protein 224 |
14 |
0.94 |
| chr10_4284967_4285185 | 0.14 |
ENSG00000207124 |
. |
272068 |
0.02 |
| chr5_54457024_54457471 | 0.14 |
GPX8 |
glutathione peroxidase 8 (putative) |
1249 |
0.31 |
| chr12_116796712_116796969 | 0.14 |
ENSG00000264037 |
. |
69283 |
0.11 |
| chr2_223912434_223912604 | 0.14 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
4013 |
0.33 |
| chr10_65479976_65480138 | 0.14 |
REEP3 |
receptor accessory protein 3 |
198934 |
0.03 |
| chr1_59478905_59479109 | 0.14 |
JUN |
jun proto-oncogene |
229222 |
0.02 |
| chr1_224805949_224806359 | 0.14 |
CNIH3 |
cornichon family AMPA receptor auxiliary protein 3 |
2159 |
0.29 |
| chr8_101555325_101555476 | 0.14 |
ANKRD46 |
ankyrin repeat domain 46 |
9847 |
0.18 |
| chr7_28944342_28944522 | 0.14 |
CTB-113D17.1 |
|
75151 |
0.1 |
| chr6_81257814_81258158 | 0.14 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
441603 |
0.01 |
| chr21_17497269_17497520 | 0.14 |
ENSG00000252273 |
. |
89565 |
0.1 |
| chr20_10720180_10720390 | 0.14 |
JAG1 |
jagged 1 |
65591 |
0.13 |
| chr3_73119964_73120214 | 0.13 |
PPP4R2 |
protein phosphatase 4, regulatory subunit 2 |
6924 |
0.14 |
| chr18_46451248_46451445 | 0.13 |
SMAD7 |
SMAD family member 7 |
23529 |
0.22 |
| chr14_27244705_27244856 | 0.13 |
NOVA1-AS1 |
NOVA1 antisense RNA 1 (head to head) |
6594 |
0.31 |
| chr3_177002301_177002503 | 0.13 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
87141 |
0.1 |
| chr10_113595251_113595472 | 0.13 |
GPAM |
glycerol-3-phosphate acyltransferase, mitochondrial |
348107 |
0.01 |
| chr14_52518827_52518978 | 0.13 |
NID2 |
nidogen 2 (osteonidogen) |
13232 |
0.2 |
| chr3_147140468_147140953 | 0.13 |
ZIC1 |
Zic family member 1 |
13539 |
0.19 |
| chr19_46920028_46920207 | 0.13 |
CCDC8 |
coiled-coil domain containing 8 |
3276 |
0.17 |
| chrX_41449282_41449446 | 0.13 |
CASK |
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
160 |
0.95 |
| chr2_199364486_199364637 | 0.13 |
ENSG00000252511 |
. |
66892 |
0.15 |
| chr13_34083543_34083799 | 0.13 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
158904 |
0.04 |
| chr3_120216529_120216680 | 0.13 |
FSTL1 |
follistatin-like 1 |
46504 |
0.16 |
| chr4_140807822_140807973 | 0.13 |
MAML3 |
mastermind-like 3 (Drosophila) |
3309 |
0.35 |
| chr9_110310779_110311295 | 0.13 |
KLF4 |
Kruppel-like factor 4 (gut) |
58274 |
0.13 |
| chr17_41622177_41622554 | 0.13 |
RP11-392O1.4 |
|
212 |
0.81 |
| chr11_65666484_65666730 | 0.13 |
FOSL1 |
FOS-like antigen 1 |
1283 |
0.23 |
| chr13_75466427_75466770 | 0.13 |
ENSG00000206812 |
. |
217126 |
0.02 |
| chr2_173037089_173037327 | 0.13 |
ENSG00000238572 |
. |
16360 |
0.23 |
| chr1_231002889_231003524 | 0.13 |
C1orf198 |
chromosome 1 open reading frame 198 |
1193 |
0.37 |
| chr4_120613511_120613662 | 0.13 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
63440 |
0.14 |
| chr12_5542106_5542552 | 0.13 |
NTF3 |
neurotrophin 3 |
1050 |
0.7 |
| chr12_12111873_12112098 | 0.13 |
ETV6 |
ets variant 6 |
73114 |
0.11 |
| chr2_181388274_181388425 | 0.13 |
ENSG00000264976 |
. |
93584 |
0.1 |
| chr4_187491209_187491867 | 0.13 |
MTNR1A |
melatonin receptor 1A |
14817 |
0.17 |
| chr5_65265501_65265702 | 0.13 |
ENSG00000252904 |
. |
8454 |
0.24 |
| chr1_92330105_92330395 | 0.13 |
TGFBR3 |
transforming growth factor, beta receptor III |
3100 |
0.29 |
| chr1_1207989_1209115 | 0.13 |
UBE2J2 |
ubiquitin-conjugating enzyme E2, J2 |
299 |
0.76 |
| chr17_79361759_79361910 | 0.13 |
RP11-1055B8.6 |
Uncharacterized protein |
7441 |
0.12 |
| chr1_1934983_1935264 | 0.13 |
C1orf222 |
chromosome 1 open reading frame 222 |
30 |
0.96 |
| chr18_7460286_7460529 | 0.13 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
106373 |
0.07 |
| chr19_45720617_45720890 | 0.13 |
EXOC3L2 |
exocyst complex component 3-like 2 |
14385 |
0.11 |
| chr11_116857809_116858077 | 0.13 |
ENSG00000264344 |
. |
28302 |
0.13 |
| chr3_117419873_117420024 | 0.13 |
ENSG00000206889 |
. |
156896 |
0.04 |
| chr5_179758220_179758506 | 0.13 |
GFPT2 |
glutamine-fructose-6-phosphate transaminase 2 |
285 |
0.91 |
| chr12_66222800_66222951 | 0.13 |
HMGA2 |
high mobility group AT-hook 2 |
3972 |
0.25 |
| chr6_152450893_152451044 | 0.13 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
38518 |
0.22 |
| chr9_111206714_111207024 | 0.13 |
ENSG00000222512 |
. |
85660 |
0.11 |
| chr6_74406883_74407034 | 0.13 |
RP11-553A21.3 |
|
1104 |
0.37 |
| chr3_130236746_130236897 | 0.13 |
COL6A6 |
collagen, type VI, alpha 6 |
42357 |
0.18 |
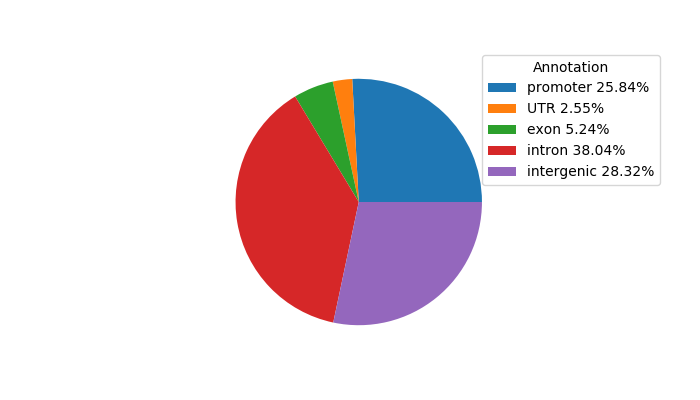
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.2 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 | 0.2 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0031223 | response to auditory stimulus(GO:0010996) auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.2 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.2 | GO:0001738 | morphogenesis of a polarized epithelium(GO:0001738) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.2 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0072071 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:2001280 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.2 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0014824 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.0 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.0 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.3 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.1 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |