Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
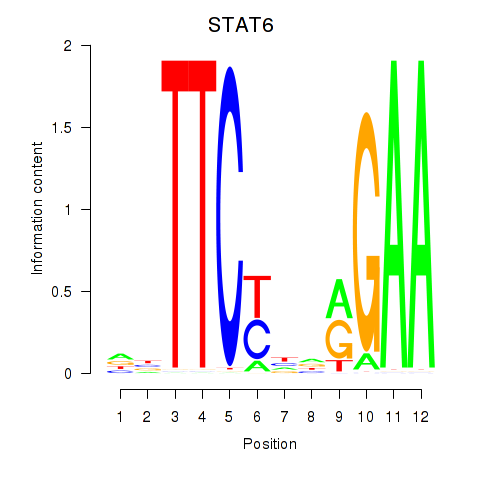
Results for STAT6
Z-value: 1.13
Transcription factors associated with STAT6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
STAT6
|
ENSG00000166888.6 | signal transducer and activator of transcription 6 |
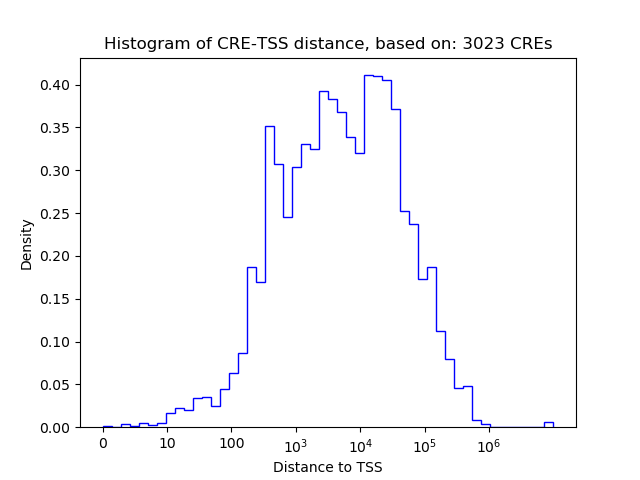
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_57525719_57525870 | STAT6 | 128 | 0.922215 | -0.72 | 2.9e-02 | Click! |
| chr12_57529685_57529883 | STAT6 | 3862 | 0.121280 | -0.68 | 4.5e-02 | Click! |
| chr12_57524952_57525103 | STAT6 | 895 | 0.387758 | -0.67 | 5.0e-02 | Click! |
| chr12_57526685_57526895 | STAT6 | 868 | 0.413961 | -0.66 | 5.3e-02 | Click! |
| chr12_57526448_57526635 | STAT6 | 619 | 0.555722 | -0.64 | 6.5e-02 | Click! |
Activity of the STAT6 motif across conditions
Conditions sorted by the z-value of the STAT6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
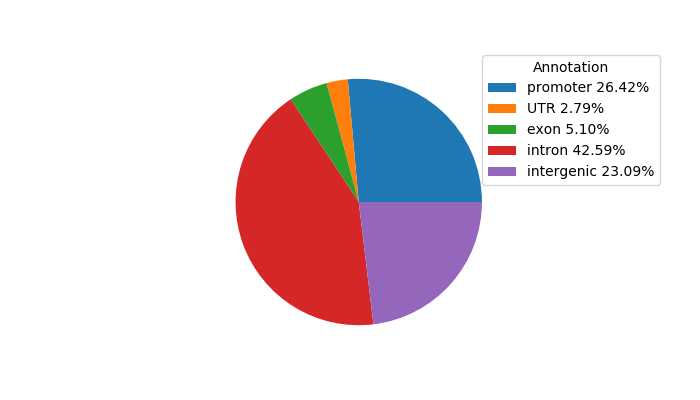
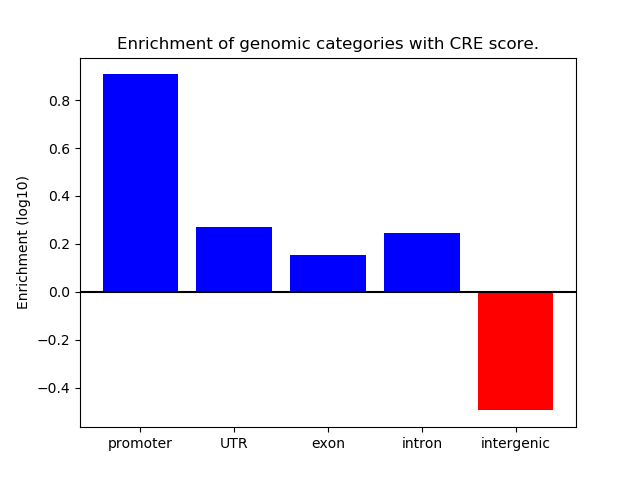
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_160766445_160766727 | 0.61 |
LY9 |
lymphocyte antigen 9 |
609 |
0.72 |
| chr14_22789744_22789895 | 0.61 |
ENSG00000251002 |
. |
111900 |
0.05 |
| chr12_47611625_47611801 | 0.53 |
PCED1B |
PC-esterase domain containing 1B |
1332 |
0.49 |
| chr1_160616915_160617313 | 0.52 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
29 |
0.97 |
| chr10_6103670_6104125 | 0.49 |
IL2RA |
interleukin 2 receptor, alpha |
356 |
0.84 |
| chr5_133452105_133452343 | 0.48 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
908 |
0.64 |
| chr22_40299734_40299885 | 0.48 |
GRAP2 |
GRB2-related adaptor protein 2 |
2696 |
0.25 |
| chr13_99957510_99957765 | 0.45 |
GPR183 |
G protein-coupled receptor 183 |
2022 |
0.34 |
| chr1_167486584_167486784 | 0.45 |
CD247 |
CD247 molecule |
1091 |
0.54 |
| chr14_99734042_99734488 | 0.44 |
BCL11B |
B-cell CLL/lymphoma 11B (zinc finger protein) |
3300 |
0.26 |
| chr8_126963197_126963348 | 0.42 |
ENSG00000206695 |
. |
50077 |
0.19 |
| chr10_73846546_73846733 | 0.41 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
1447 |
0.48 |
| chr6_144665292_144666004 | 0.37 |
UTRN |
utrophin |
411 |
0.88 |
| chr14_99726303_99726510 | 0.37 |
AL109767.1 |
|
2879 |
0.29 |
| chr3_73126790_73126941 | 0.36 |
ENSG00000238416 |
. |
4273 |
0.16 |
| chr17_33566033_33566184 | 0.36 |
RP11-799D4.4 |
|
1108 |
0.42 |
| chr17_33568427_33568649 | 0.35 |
RP11-799D4.4 |
|
1444 |
0.28 |
| chr11_2322790_2323148 | 0.35 |
C11orf21 |
chromosome 11 open reading frame 21 |
174 |
0.62 |
| chr8_134069781_134069970 | 0.35 |
SLA |
Src-like-adaptor |
2728 |
0.34 |
| chr18_67623543_67623764 | 0.33 |
CD226 |
CD226 molecule |
253 |
0.95 |
| chr8_134084186_134084496 | 0.33 |
SLA |
Src-like-adaptor |
11738 |
0.24 |
| chr12_6555099_6555250 | 0.33 |
CD27 |
CD27 molecule |
1141 |
0.3 |
| chr7_3083802_3083953 | 0.33 |
CARD11 |
caspase recruitment domain family, member 11 |
298 |
0.93 |
| chr8_38219852_38220003 | 0.33 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
18412 |
0.12 |
| chr6_93898534_93898685 | 0.33 |
EPHA7 |
EPH receptor A7 |
230635 |
0.02 |
| chr1_192545263_192545458 | 0.32 |
RGS1 |
regulator of G-protein signaling 1 |
457 |
0.84 |
| chr18_2982488_2982849 | 0.32 |
LPIN2 |
lipin 2 |
203 |
0.93 |
| chr7_150148744_150149359 | 0.32 |
GIMAP8 |
GTPase, IMAP family member 8 |
1333 |
0.41 |
| chrY_16636977_16637245 | 0.31 |
NLGN4Y |
neuroligin 4, Y-linked |
657 |
0.84 |
| chr6_122729994_122730284 | 0.31 |
HSF2 |
heat shock transcription factor 2 |
9443 |
0.26 |
| chr4_109084692_109084886 | 0.31 |
LEF1 |
lymphoid enhancer-binding factor 1 |
2668 |
0.31 |
| chr7_1086655_1086891 | 0.30 |
GPR146 |
G protein-coupled receptor 146 |
2561 |
0.15 |
| chr11_413234_413456 | 0.30 |
SIGIRR |
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
1603 |
0.21 |
| chr2_106473982_106474172 | 0.30 |
AC009505.2 |
|
444 |
0.86 |
| chr14_99708890_99709242 | 0.29 |
AL109767.1 |
|
20219 |
0.2 |
| chr6_24930869_24931159 | 0.29 |
FAM65B |
family with sequence similarity 65, member B |
5174 |
0.26 |
| chr9_136006193_136006379 | 0.29 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
218 |
0.91 |
| chr18_2981451_2981852 | 0.29 |
LPIN2 |
lipin 2 |
1220 |
0.42 |
| chr10_100503637_100503788 | 0.28 |
HPS1 |
Hermansky-Pudlak syndrome 1 |
297029 |
0.01 |
| chr13_42848803_42848954 | 0.28 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
2589 |
0.4 |
| chr5_118693818_118693969 | 0.28 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2163 |
0.35 |
| chr13_52516667_52516944 | 0.28 |
ATP7B |
ATPase, Cu++ transporting, beta polypeptide |
19246 |
0.2 |
| chr5_130617597_130618118 | 0.28 |
CDC42SE2 |
CDC42 small effector 2 |
18064 |
0.27 |
| chr18_60819434_60819585 | 0.27 |
RP11-299P2.1 |
|
956 |
0.65 |
| chr14_35399812_35400018 | 0.27 |
ENSG00000199980 |
. |
2834 |
0.24 |
| chr16_78776576_78776802 | 0.27 |
RP11-319G9.3 |
|
154373 |
0.04 |
| chr12_109026876_109027027 | 0.27 |
SELPLG |
selectin P ligand |
719 |
0.55 |
| chr10_127686444_127686595 | 0.27 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
2533 |
0.31 |
| chr6_35267244_35267414 | 0.27 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
1700 |
0.35 |
| chr14_100539294_100539636 | 0.26 |
CTD-2376I20.1 |
|
1750 |
0.29 |
| chr8_8203535_8203805 | 0.26 |
SGK223 |
Tyrosine-protein kinase SgK223 |
35587 |
0.18 |
| chr4_122103035_122103186 | 0.26 |
ENSG00000252183 |
. |
10948 |
0.22 |
| chrY_21906454_21906814 | 0.26 |
KDM5D |
lysine (K)-specific demethylase 5D |
13 |
0.99 |
| chr1_241715487_241715755 | 0.26 |
KMO |
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
16314 |
0.2 |
| chr2_109605420_109605703 | 0.26 |
EDAR |
ectodysplasin A receptor |
164 |
0.97 |
| chr12_9819897_9820101 | 0.26 |
CLEC2D |
C-type lectin domain family 2, member D |
2310 |
0.2 |
| chr14_99727366_99727576 | 0.26 |
AL109767.1 |
|
1814 |
0.39 |
| chr3_69249720_69249913 | 0.26 |
FRMD4B |
FERM domain containing 4B |
183 |
0.96 |
| chr11_46352507_46352658 | 0.26 |
DGKZ |
diacylglycerol kinase, zeta |
1873 |
0.29 |
| chr9_114836879_114837030 | 0.26 |
RP11-4O1.2 |
|
36944 |
0.14 |
| chr1_111214730_111214881 | 0.26 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
2850 |
0.25 |
| chr4_26398560_26398711 | 0.26 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
31707 |
0.23 |
| chr9_95727852_95728104 | 0.25 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1735 |
0.41 |
| chr11_66795405_66795556 | 0.25 |
SYT12 |
synaptotagmin XII |
449 |
0.77 |
| chr4_40195304_40195587 | 0.25 |
RHOH |
ras homolog family member H |
162 |
0.96 |
| chr14_22974481_22974956 | 0.25 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18547 |
0.09 |
| chr3_46396239_46396534 | 0.25 |
CCR2 |
chemokine (C-C motif) receptor 2 |
769 |
0.62 |
| chr1_186291714_186291865 | 0.25 |
ENSG00000202025 |
. |
10729 |
0.17 |
| chr1_154300198_154300512 | 0.24 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
79 |
0.94 |
| chr10_62492315_62493446 | 0.24 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
368 |
0.92 |
| chr5_35857058_35857643 | 0.24 |
IL7R |
interleukin 7 receptor |
356 |
0.89 |
| chr3_151973155_151973306 | 0.24 |
MBNL1 |
muscleblind-like splicing regulator 1 |
12599 |
0.22 |
| chr7_90742755_90742906 | 0.24 |
FZD1 |
frizzled family receptor 1 |
150953 |
0.05 |
| chr2_161994699_161994850 | 0.24 |
TANK |
TRAF family member-associated NFKB activator |
1308 |
0.52 |
| chr14_102280065_102280216 | 0.24 |
CTD-2017C7.2 |
|
3482 |
0.18 |
| chr7_107045766_107045917 | 0.24 |
GPR22 |
G protein-coupled receptor 22 |
64664 |
0.11 |
| chr9_78639191_78639342 | 0.24 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
71627 |
0.13 |
| chr2_68961577_68961872 | 0.24 |
ARHGAP25 |
Rho GTPase activating protein 25 |
189 |
0.96 |
| chr9_130666902_130667485 | 0.24 |
ST6GALNAC6 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6 |
416 |
0.68 |
| chr21_19165639_19165946 | 0.23 |
AL109761.5 |
|
13 |
0.98 |
| chr13_49391256_49391452 | 0.23 |
ENSG00000222391 |
. |
65062 |
0.12 |
| chr4_36244120_36244417 | 0.23 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
1293 |
0.39 |
| chr17_41738444_41738624 | 0.23 |
MEOX1 |
mesenchyme homeobox 1 |
397 |
0.85 |
| chr8_22552129_22552945 | 0.23 |
EGR3 |
early growth response 3 |
1722 |
0.26 |
| chr1_153513589_153513800 | 0.23 |
S100A5 |
S100 calcium binding protein A5 |
524 |
0.57 |
| chr17_6949635_6950042 | 0.23 |
SLC16A11 |
solute carrier family 16, member 11 |
2596 |
0.11 |
| chr3_156201429_156202178 | 0.23 |
KCNAB1-AS1 |
KCNAB1 antisense RNA 1 |
37109 |
0.17 |
| chr13_23174183_23174334 | 0.23 |
ENSG00000253094 |
. |
203026 |
0.03 |
| chr16_26606103_26606254 | 0.23 |
ENSG00000264935 |
. |
141372 |
0.05 |
| chr2_225811162_225811313 | 0.22 |
DOCK10 |
dedicator of cytokinesis 10 |
545 |
0.86 |
| chr15_78361308_78361764 | 0.22 |
TBC1D2B |
TBC1 domain family, member 2B |
3071 |
0.18 |
| chr6_18348172_18348323 | 0.22 |
RNF144B |
ring finger protein 144B |
20532 |
0.18 |
| chr10_32748199_32748350 | 0.22 |
CCDC7 |
coiled-coil domain containing 7 |
7725 |
0.23 |
| chr11_128700598_128700752 | 0.22 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
11754 |
0.2 |
| chr1_169679248_169679951 | 0.22 |
SELL |
selectin L |
1240 |
0.48 |
| chr8_80268950_80269101 | 0.22 |
STMN2 |
stathmin-like 2 |
254024 |
0.02 |
| chr3_43736623_43736774 | 0.22 |
ANO10 |
anoctamin 10 |
3612 |
0.27 |
| chr11_14097283_14097490 | 0.21 |
ENSG00000212365 |
. |
59146 |
0.15 |
| chr6_159461701_159461911 | 0.21 |
TAGAP |
T-cell activation RhoGTPase activating protein |
4244 |
0.22 |
| chr11_6764514_6764763 | 0.21 |
GVINP1 |
GTPase, very large interferon inducible pseudogene 1 |
21527 |
0.11 |
| chr4_38513548_38513699 | 0.21 |
RP11-617D20.1 |
|
112573 |
0.06 |
| chr4_70625874_70626025 | 0.21 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
367 |
0.91 |
| chr13_48894505_48894712 | 0.21 |
RB1 |
retinoblastoma 1 |
16697 |
0.25 |
| chr5_39197523_39197702 | 0.21 |
FYB |
FYN binding protein |
5517 |
0.3 |
| chr6_45982173_45982324 | 0.21 |
CLIC5 |
chloride intracellular channel 5 |
1317 |
0.48 |
| chr4_187639922_187640073 | 0.21 |
FAT1 |
FAT atypical cadherin 1 |
5012 |
0.32 |
| chr7_138788632_138788799 | 0.20 |
ZC3HAV1 |
zinc finger CCCH-type, antiviral 1 |
5385 |
0.23 |
| chr3_5178219_5178370 | 0.20 |
ARL8B |
ADP-ribosylation factor-like 8B |
14313 |
0.17 |
| chr14_50448972_50449232 | 0.20 |
C14orf182 |
chromosome 14 open reading frame 182 |
25136 |
0.15 |
| chr14_55852967_55853226 | 0.20 |
ATG14 |
autophagy related 14 |
25480 |
0.18 |
| chr15_38855839_38856970 | 0.20 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
432 |
0.84 |
| chr8_128987632_128988045 | 0.20 |
ENSG00000221771 |
. |
14959 |
0.16 |
| chr13_40532608_40532878 | 0.20 |
ENSG00000212553 |
. |
101379 |
0.08 |
| chr5_40684769_40685057 | 0.20 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
5313 |
0.23 |
| chr21_38352575_38353327 | 0.20 |
HLCS |
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
23 |
0.97 |
| chr3_128838297_128838517 | 0.20 |
RAB43 |
RAB43, member RAS oncogene family |
2217 |
0.2 |
| chr10_105453143_105453834 | 0.20 |
SH3PXD2A |
SH3 and PX domains 2A |
558 |
0.78 |
| chr1_117515195_117515346 | 0.20 |
ENSG00000252510 |
. |
10177 |
0.17 |
| chr8_59352943_59353094 | 0.20 |
UBXN2B |
UBX domain protein 2B |
23529 |
0.23 |
| chr4_150907034_150907185 | 0.20 |
ENSG00000207121 |
. |
51581 |
0.16 |
| chrY_16635792_16636017 | 0.20 |
NLGN4Y |
neuroligin 4, Y-linked |
278 |
0.96 |
| chr1_62905567_62905816 | 0.19 |
USP1 |
ubiquitin specific peptidase 1 |
3365 |
0.33 |
| chr2_235359845_235359996 | 0.19 |
ARL4C |
ADP-ribosylation factor-like 4C |
45324 |
0.21 |
| chr6_117870110_117870478 | 0.19 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
53233 |
0.12 |
| chr13_97877051_97877292 | 0.19 |
MBNL2 |
muscleblind-like splicing regulator 2 |
2562 |
0.41 |
| chr13_99992644_99992897 | 0.19 |
ENSG00000207719 |
. |
15615 |
0.19 |
| chr5_130588828_130589214 | 0.19 |
CDC42SE2 |
CDC42 small effector 2 |
10681 |
0.28 |
| chr2_114652613_114652764 | 0.19 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
4513 |
0.26 |
| chr10_15296464_15296906 | 0.19 |
RP11-25G10.2 |
|
17194 |
0.24 |
| chr22_40858602_40858925 | 0.19 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
659 |
0.7 |
| chrY_15815919_15816489 | 0.19 |
TMSB4Y |
thymosin beta 4, Y-linked |
757 |
0.72 |
| chr3_128835091_128835242 | 0.19 |
RAB43 |
RAB43, member RAS oncogene family |
5458 |
0.13 |
| chrX_64893704_64894104 | 0.19 |
MSN |
moesin |
6367 |
0.32 |
| chr5_39207439_39207590 | 0.19 |
FYB |
FYN binding protein |
4385 |
0.32 |
| chr12_40022874_40023025 | 0.19 |
C12orf40 |
chromosome 12 open reading frame 40 |
2964 |
0.31 |
| chr5_110565458_110565614 | 0.19 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
5752 |
0.25 |
| chr4_122103316_122103738 | 0.18 |
ENSG00000252183 |
. |
10531 |
0.22 |
| chr2_238609518_238609709 | 0.18 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
8631 |
0.24 |
| chr22_33278140_33278291 | 0.18 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
80528 |
0.1 |
| chr9_132890907_132891166 | 0.18 |
ENSG00000223188 |
. |
7951 |
0.19 |
| chr6_109694670_109694821 | 0.18 |
CD164 |
CD164 molecule, sialomucin |
8158 |
0.15 |
| chr4_40230569_40230720 | 0.18 |
RHOH |
ras homolog family member H |
28680 |
0.18 |
| chr7_69471547_69471698 | 0.18 |
AUTS2 |
autism susceptibility candidate 2 |
407025 |
0.01 |
| chr13_41578397_41578548 | 0.18 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
14978 |
0.19 |
| chrX_46436832_46437001 | 0.18 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
3697 |
0.27 |
| chr7_43626236_43626525 | 0.18 |
STK17A |
serine/threonine kinase 17a |
3716 |
0.23 |
| chr1_4558588_4558739 | 0.18 |
AJAP1 |
adherens junctions associated protein 1 |
156129 |
0.04 |
| chr22_33040985_33041136 | 0.18 |
ENSG00000251890 |
. |
9876 |
0.25 |
| chr4_40213524_40213675 | 0.18 |
RHOH |
ras homolog family member H |
11635 |
0.22 |
| chr21_42798492_42799079 | 0.18 |
MX1 |
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
617 |
0.73 |
| chrX_12991303_12991531 | 0.18 |
TMSB4X |
thymosin beta 4, X-linked |
1810 |
0.41 |
| chr1_92846684_92846835 | 0.18 |
ENSG00000242764 |
. |
21483 |
0.21 |
| chr5_175080247_175080498 | 0.18 |
HRH2 |
histamine receptor H2 |
4661 |
0.24 |
| chr2_9922535_9922686 | 0.18 |
ENSG00000200034 |
. |
41698 |
0.16 |
| chr17_38721330_38721617 | 0.18 |
CCR7 |
chemokine (C-C motif) receptor 7 |
238 |
0.92 |
| chr2_20776643_20776868 | 0.18 |
HS1BP3-IT1 |
HS1BP3 intronic transcript 1 (non-protein coding) |
15553 |
0.21 |
| chr2_27485345_27486489 | 0.18 |
SLC30A3 |
solute carrier family 30 (zinc transporter), member 3 |
178 |
0.89 |
| chr14_90147435_90147962 | 0.17 |
ENSG00000252655 |
. |
30451 |
0.14 |
| chrX_48542477_48542733 | 0.17 |
WAS |
Wiskott-Aldrich syndrome |
437 |
0.75 |
| chr1_7999953_8000204 | 0.17 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
848 |
0.6 |
| chr17_79025867_79026018 | 0.17 |
BAIAP2 |
BAI1-associated protein 2 |
1220 |
0.37 |
| chr6_10529624_10529892 | 0.17 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
1169 |
0.49 |
| chr5_159903210_159903361 | 0.17 |
ENSG00000265237 |
. |
1876 |
0.31 |
| chr8_37751638_37751789 | 0.17 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
5259 |
0.15 |
| chrX_103357104_103358067 | 0.17 |
ZCCHC18 |
zinc finger, CCHC domain containing 18 |
383 |
0.86 |
| chr1_9713063_9713437 | 0.17 |
C1orf200 |
chromosome 1 open reading frame 200 |
1394 |
0.31 |
| chr10_121965154_121965305 | 0.17 |
ENSG00000221402 |
. |
81491 |
0.11 |
| chrY_7542080_7542231 | 0.17 |
RFTN1P1 |
raftlin, lipid raft linker 1 pseudogene 1 |
102593 |
0.08 |
| chr3_16329082_16329264 | 0.17 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
18425 |
0.15 |
| chr7_102489527_102489678 | 0.17 |
ENSG00000238324 |
. |
8295 |
0.18 |
| chr3_171852368_171852519 | 0.17 |
FNDC3B |
fibronectin type III domain containing 3B |
7679 |
0.29 |
| chr12_88222507_88222658 | 0.17 |
C12orf50 |
chromosome 12 open reading frame 50 |
199158 |
0.03 |
| chr11_88087417_88087693 | 0.17 |
CTSC |
cathepsin C |
16600 |
0.28 |
| chr6_144881069_144881220 | 0.17 |
UTRN |
utrophin |
23202 |
0.28 |
| chr12_92899150_92899489 | 0.17 |
ENSG00000238865 |
. |
39444 |
0.17 |
| chr2_30459993_30460144 | 0.17 |
LBH |
limb bud and heart development |
5022 |
0.26 |
| chr3_63338970_63339121 | 0.16 |
ENSG00000252662 |
. |
18582 |
0.26 |
| chr17_33864237_33864745 | 0.16 |
SLFN12L |
schlafen family member 12-like |
389 |
0.76 |
| chr14_82916567_82916718 | 0.16 |
ENSG00000238978 |
. |
11790 |
0.33 |
| chr11_113730700_113730851 | 0.16 |
ENSG00000201687 |
. |
707 |
0.64 |
| chr3_177314905_177315095 | 0.16 |
ENSG00000200288 |
. |
27197 |
0.25 |
| chr1_47109278_47109429 | 0.16 |
ATPAF1 |
ATP synthase mitochondrial F1 complex assembly factor 1 |
1575 |
0.3 |
| chr10_33415014_33415165 | 0.16 |
ENSG00000263576 |
. |
27525 |
0.2 |
| chr2_29121252_29121403 | 0.16 |
WDR43 |
WD repeat domain 43 |
2657 |
0.17 |
| chr10_5752759_5752996 | 0.16 |
RP11-336A10.2 |
|
3509 |
0.22 |
| chr1_167059294_167059481 | 0.16 |
GPA33 |
glycoprotein A33 (transmembrane) |
481 |
0.77 |
| chr2_85830286_85830577 | 0.16 |
TMEM150A |
transmembrane protein 150A |
610 |
0.53 |
| chr1_40848541_40848692 | 0.16 |
SMAP2 |
small ArfGAP2 |
8296 |
0.18 |
| chr1_165393515_165393674 | 0.16 |
RXRG |
retinoid X receptor, gamma |
20839 |
0.21 |
| chr14_55598726_55598877 | 0.16 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
2823 |
0.31 |
| chr2_107128169_107128320 | 0.16 |
CD8BP |
CD8b molecule pseudogene |
24361 |
0.18 |
| chrX_106070087_106070238 | 0.15 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
8535 |
0.25 |
| chr1_9473851_9474134 | 0.15 |
ENSG00000252956 |
. |
23845 |
0.2 |
| chr1_108211265_108211416 | 0.15 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
19786 |
0.27 |
| chr19_17862777_17862928 | 0.15 |
FCHO1 |
FCH domain only 1 |
507 |
0.71 |
| chr7_105902270_105902421 | 0.15 |
NAMPT |
nicotinamide phosphoribosyltransferase |
23022 |
0.19 |
| chr16_74732357_74732554 | 0.15 |
MLKL |
mixed lineage kinase domain-like |
2233 |
0.3 |
| chr2_30458431_30458703 | 0.15 |
LBH |
limb bud and heart development |
3521 |
0.29 |
| chr17_39810938_39811089 | 0.15 |
KRT42P |
keratin 42 pseudogene |
24169 |
0.09 |
| chr6_109803101_109803270 | 0.15 |
ZBTB24 |
zinc finger and BTB domain containing 24 |
1255 |
0.32 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.1 | 0.4 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.1 | 0.2 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 0.4 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.1 | 0.3 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.3 | GO:0002855 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.1 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.1 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:0070602 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.5 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.2 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.3 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.0 | 0.1 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0021612 | cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0045683 | negative regulation of epidermis development(GO:0045683) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.0 | GO:1900120 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.0 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0042584 | chromaffin granule(GO:0042583) chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.7 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |