Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
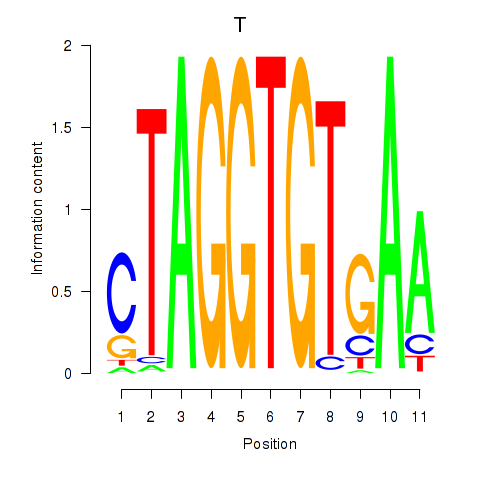
Results for T
Z-value: 0.61
Transcription factors associated with T
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
T
|
ENSG00000164458.5 | T |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_166582350_166582501 | T | 237 | 0.943451 | -0.44 | 2.3e-01 | Click! |
| chr6_166581874_166582172 | T | 84 | 0.977763 | 0.03 | 9.3e-01 | Click! |
Activity of the T motif across conditions
Conditions sorted by the z-value of the T motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
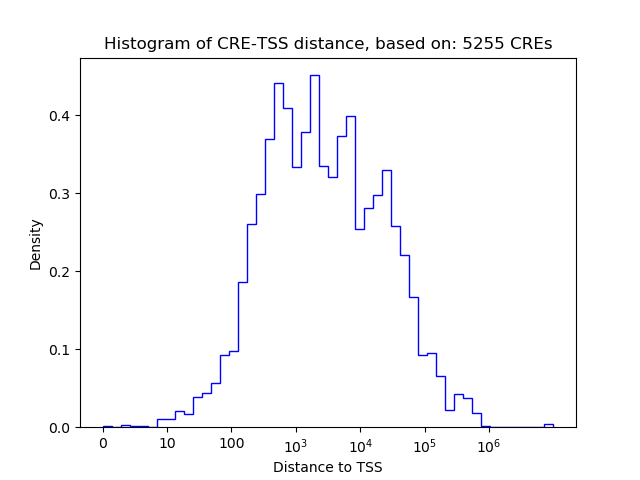
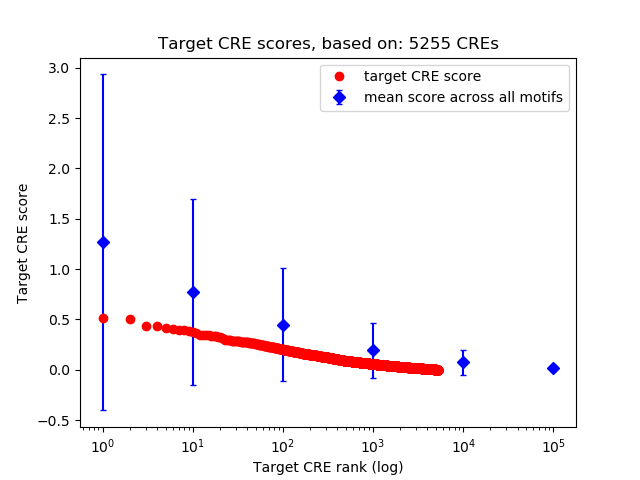
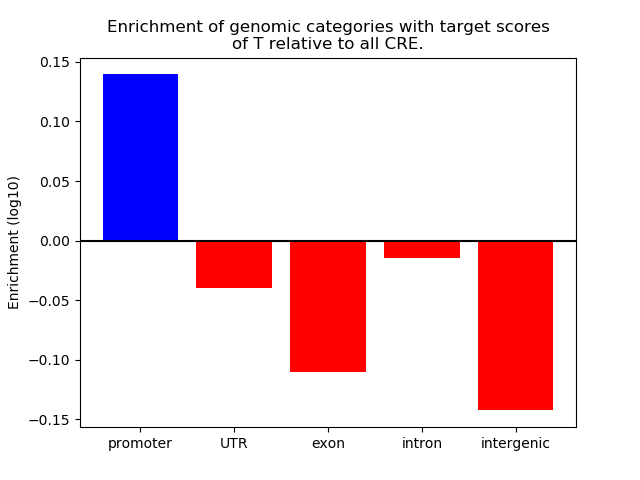
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_120687736_120687954 | 0.52 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
493 |
0.83 |
| chr1_203735045_203735272 | 0.50 |
LAX1 |
lymphocyte transmembrane adaptor 1 |
853 |
0.57 |
| chr15_40599321_40599893 | 0.44 |
PLCB2 |
phospholipase C, beta 2 |
419 |
0.68 |
| chr10_6103505_6103656 | 0.43 |
IL2RA |
interleukin 2 receptor, alpha |
673 |
0.64 |
| chr3_114014633_114014828 | 0.42 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
896 |
0.58 |
| chrX_128913998_128914362 | 0.41 |
SASH3 |
SAM and SH3 domain containing 3 |
220 |
0.94 |
| chr14_92331424_92331867 | 0.40 |
TC2N |
tandem C2 domains, nuclear |
2228 |
0.32 |
| chr9_95727676_95727827 | 0.40 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1508 |
0.45 |
| chr21_15917916_15918619 | 0.39 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr3_150920478_150920863 | 0.38 |
GPR171 |
G protein-coupled receptor 171 |
309 |
0.87 |
| chr9_95728150_95728303 | 0.36 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1983 |
0.37 |
| chr13_99957510_99957765 | 0.35 |
GPR183 |
G protein-coupled receptor 183 |
2022 |
0.34 |
| chr4_100738344_100738640 | 0.35 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
489 |
0.85 |
| chr1_6525547_6525781 | 0.35 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
26 |
0.96 |
| chr1_9689695_9689868 | 0.34 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
22009 |
0.15 |
| chr1_198615772_198616039 | 0.34 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
7613 |
0.25 |
| chr7_962027_962293 | 0.33 |
ADAP1 |
ArfGAP with dual PH domains 1 |
1634 |
0.31 |
| chr6_130536667_130536818 | 0.33 |
SAMD3 |
sterile alpha motif domain containing 3 |
45 |
0.99 |
| chr3_111261315_111261845 | 0.33 |
CD96 |
CD96 molecule |
583 |
0.82 |
| chr8_48515171_48515322 | 0.32 |
SPIDR |
scaffolding protein involved in DNA repair |
56980 |
0.12 |
| chr6_20336872_20337023 | 0.31 |
E2F3 |
E2F transcription factor 3 |
65451 |
0.1 |
| chr2_2650168_2650319 | 0.31 |
MYT1L |
myelin transcription factor 1-like |
315277 |
0.01 |
| chr14_102280520_102280788 | 0.30 |
CTD-2017C7.2 |
|
3996 |
0.17 |
| chr14_22999291_22999566 | 0.30 |
TRAJ15 |
T cell receptor alpha joining 15 |
848 |
0.46 |
| chr14_22994017_22994177 | 0.29 |
TRAJ15 |
T cell receptor alpha joining 15 |
4483 |
0.13 |
| chr3_46396063_46396214 | 0.29 |
CCR2 |
chemokine (C-C motif) receptor 2 |
521 |
0.76 |
| chr2_120690048_120690199 | 0.29 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
71 |
0.98 |
| chr11_118177311_118177735 | 0.29 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
1909 |
0.24 |
| chr22_20784096_20784540 | 0.29 |
SCARF2 |
scavenger receptor class F, member 2 |
7794 |
0.11 |
| chr1_198616268_198616512 | 0.29 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
8098 |
0.25 |
| chr5_150593261_150593549 | 0.29 |
GM2A |
GM2 ganglioside activator |
1694 |
0.36 |
| chr5_180231596_180231747 | 0.28 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
748 |
0.63 |
| chr16_88717586_88717845 | 0.28 |
CYBA |
cytochrome b-245, alpha polypeptide |
155 |
0.9 |
| chr20_45623189_45623371 | 0.28 |
EYA2 |
eyes absent homolog 2 (Drosophila) |
4640 |
0.32 |
| chr14_55800013_55800175 | 0.28 |
RP11-665C16.5 |
|
6125 |
0.25 |
| chr11_2466894_2467127 | 0.28 |
KCNQ1 |
potassium voltage-gated channel, KQT-like subfamily, member 1 |
789 |
0.56 |
| chr11_127906960_127907260 | 0.27 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
468179 |
0.01 |
| chr13_41589998_41590808 | 0.27 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
3047 |
0.26 |
| chr22_40858929_40859404 | 0.27 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
256 |
0.92 |
| chr3_185331003_185331154 | 0.27 |
SENP2 |
SUMO1/sentrin/SMT3 specific peptidase 2 |
65 |
0.98 |
| chr17_72735282_72735631 | 0.27 |
RAB37 |
RAB37, member RAS oncogene family |
2085 |
0.18 |
| chr2_237446743_237446920 | 0.27 |
ACKR3 |
atypical chemokine receptor 3 |
29599 |
0.19 |
| chr13_46749322_46749478 | 0.27 |
ENSG00000240767 |
. |
5517 |
0.17 |
| chr2_235400500_235400697 | 0.27 |
ARL4C |
ADP-ribosylation factor-like 4C |
4646 |
0.36 |
| chr19_13215909_13216097 | 0.27 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
2028 |
0.18 |
| chr6_38019767_38019918 | 0.26 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
122107 |
0.06 |
| chr22_40299734_40299885 | 0.26 |
GRAP2 |
GRB2-related adaptor protein 2 |
2696 |
0.25 |
| chr12_32119532_32119743 | 0.26 |
KIAA1551 |
KIAA1551 |
4214 |
0.28 |
| chr22_40307152_40307436 | 0.26 |
GRAP2 |
GRB2-related adaptor protein 2 |
10181 |
0.17 |
| chr16_53491555_53491772 | 0.26 |
RBL2 |
retinoblastoma-like 2 (p130) |
7675 |
0.16 |
| chr1_67398755_67399224 | 0.26 |
MIER1 |
mesoderm induction early response 1, transcriptional regulator |
3063 |
0.27 |
| chrX_40030247_40030398 | 0.26 |
BCOR |
BCL6 corepressor |
6251 |
0.32 |
| chr1_12230241_12230523 | 0.26 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
3322 |
0.2 |
| chr16_84634203_84634572 | 0.25 |
RP11-61F12.1 |
|
6388 |
0.18 |
| chr22_40297106_40297391 | 0.25 |
GRAP2 |
GRB2-related adaptor protein 2 |
135 |
0.96 |
| chr14_55799126_55799413 | 0.25 |
RP11-665C16.5 |
|
6950 |
0.25 |
| chr4_38916539_38916690 | 0.25 |
ENSG00000207944 |
. |
46961 |
0.11 |
| chr12_46062713_46062864 | 0.24 |
ARID2 |
AT rich interactive domain 2 (ARID, RFX-like) |
60660 |
0.15 |
| chr15_65597552_65597703 | 0.24 |
ENSG00000200156 |
. |
612 |
0.64 |
| chr12_1920714_1920865 | 0.24 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
97 |
0.97 |
| chr10_71332813_71333013 | 0.24 |
NEUROG3 |
neurogenin 3 |
81 |
0.97 |
| chr5_32583768_32584065 | 0.24 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
1689 |
0.49 |
| chr5_32532091_32532542 | 0.24 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
392 |
0.9 |
| chr6_12061250_12061691 | 0.24 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
45650 |
0.18 |
| chrX_57313067_57313960 | 0.24 |
FAAH2 |
fatty acid amide hydrolase 2 |
374 |
0.92 |
| chr5_59062656_59062927 | 0.23 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
1671 |
0.51 |
| chr15_60296272_60296423 | 0.23 |
FOXB1 |
forkhead box B1 |
74 |
0.99 |
| chr18_60982497_60982692 | 0.23 |
RP11-28F1.2 |
|
1279 |
0.41 |
| chr14_98444010_98444505 | 0.23 |
C14orf64 |
chromosome 14 open reading frame 64 |
126 |
0.98 |
| chr19_49840027_49840178 | 0.23 |
CD37 |
CD37 molecule |
370 |
0.72 |
| chr2_112178355_112178506 | 0.23 |
ENSG00000266139 |
. |
99762 |
0.09 |
| chr22_40640074_40640407 | 0.23 |
TNRC6B |
trinucleotide repeat containing 6B |
20767 |
0.22 |
| chr10_127731141_127731292 | 0.23 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
47230 |
0.14 |
| chr7_35730534_35730808 | 0.23 |
HERPUD2 |
HERPUD family member 2 |
3505 |
0.27 |
| chr2_101916869_101917041 | 0.23 |
RNF149 |
ring finger protein 149 |
8203 |
0.17 |
| chr11_45377791_45377942 | 0.23 |
SYT13 |
synaptotagmin XIII |
69996 |
0.11 |
| chr4_14169430_14169581 | 0.22 |
ENSG00000252092 |
. |
509254 |
0.0 |
| chrX_128914480_128914899 | 0.22 |
SASH3 |
SAM and SH3 domain containing 3 |
729 |
0.68 |
| chr19_14260226_14260973 | 0.22 |
LPHN1 |
latrophilin 1 |
3052 |
0.14 |
| chr12_9911780_9911931 | 0.22 |
CD69 |
CD69 molecule |
1642 |
0.33 |
| chr10_102270947_102271098 | 0.22 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
8569 |
0.16 |
| chr1_150535533_150535831 | 0.22 |
ADAMTSL4-AS1 |
ADAMTSL4 antisense RNA 1 |
1713 |
0.17 |
| chr1_167485802_167485953 | 0.22 |
CD247 |
CD247 molecule |
1898 |
0.36 |
| chr10_9271793_9271944 | 0.22 |
ENSG00000212505 |
. |
573074 |
0.0 |
| chr1_200991644_200992373 | 0.22 |
KIF21B |
kinesin family member 21B |
528 |
0.78 |
| chr4_6912691_6912996 | 0.21 |
TBC1D14 |
TBC1 domain family, member 14 |
868 |
0.61 |
| chr1_244483301_244483484 | 0.21 |
C1orf100 |
chromosome 1 open reading frame 100 |
32545 |
0.2 |
| chrX_70323668_70324217 | 0.21 |
CXorf65 |
chromosome X open reading frame 65 |
2513 |
0.16 |
| chr2_225808771_225808936 | 0.21 |
DOCK10 |
dedicator of cytokinesis 10 |
2929 |
0.39 |
| chr10_6627252_6627403 | 0.21 |
PRKCQ |
protein kinase C, theta |
5064 |
0.34 |
| chr17_76730952_76731103 | 0.21 |
CYTH1 |
cytohesin 1 |
1945 |
0.35 |
| chr5_67576190_67576341 | 0.21 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
130 |
0.98 |
| chr8_27183037_27183778 | 0.21 |
PTK2B |
protein tyrosine kinase 2 beta |
326 |
0.9 |
| chr20_62271176_62271486 | 0.21 |
CTD-3184A7.4 |
|
12728 |
0.09 |
| chr16_31468522_31468673 | 0.20 |
ARMC5 |
armadillo repeat containing 5 |
804 |
0.4 |
| chr1_27686628_27686822 | 0.20 |
MAP3K6 |
mitogen-activated protein kinase kinase kinase 6 |
3717 |
0.14 |
| chr17_2699201_2699651 | 0.20 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
306 |
0.89 |
| chr19_16477276_16478284 | 0.20 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
5016 |
0.17 |
| chr15_60882704_60882968 | 0.20 |
RORA |
RAR-related orphan receptor A |
1904 |
0.39 |
| chr1_6054566_6054852 | 0.20 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
1938 |
0.28 |
| chr17_20602116_20602267 | 0.20 |
AC126365.1 |
|
17605 |
0.23 |
| chr2_68994473_68994840 | 0.20 |
ARHGAP25 |
Rho GTPase activating protein 25 |
7277 |
0.25 |
| chr2_109238478_109238884 | 0.20 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
959 |
0.64 |
| chr19_30158859_30159039 | 0.20 |
PLEKHF1 |
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
1168 |
0.55 |
| chr10_23631330_23631481 | 0.20 |
RP11-371A19.2 |
|
1481 |
0.31 |
| chr1_89740013_89740507 | 0.20 |
GBP5 |
guanylate binding protein 5 |
1716 |
0.37 |
| chr1_111173785_111173949 | 0.20 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
229 |
0.93 |
| chr10_102133099_102133250 | 0.20 |
ENSG00000212325 |
. |
25029 |
0.12 |
| chr17_56410108_56410593 | 0.20 |
MIR142 |
microRNA 142 |
481 |
0.66 |
| chr3_71772714_71772865 | 0.20 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
1737 |
0.4 |
| chr8_38215592_38215743 | 0.20 |
RP11-513D5.2 |
|
22168 |
0.11 |
| chr19_6773186_6773436 | 0.19 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
342 |
0.82 |
| chr1_19284092_19284243 | 0.19 |
IFFO2 |
intermediate filament family orphan 2 |
987 |
0.58 |
| chr12_12712693_12713134 | 0.19 |
DUSP16 |
dual specificity phosphatase 16 |
1098 |
0.58 |
| chr14_22236992_22237143 | 0.19 |
ENSG00000222776 |
. |
11718 |
0.25 |
| chr2_70321532_70321889 | 0.19 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
5732 |
0.17 |
| chr6_167367959_167368110 | 0.19 |
RP11-514O12.4 |
|
1578 |
0.28 |
| chrX_144503620_144503771 | 0.19 |
SPANXN1 |
SPANX family, member N1 |
175347 |
0.03 |
| chr6_13428686_13428841 | 0.19 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
20394 |
0.18 |
| chr1_161008159_161008677 | 0.19 |
TSTD1 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
101 |
0.93 |
| chr7_8010063_8010681 | 0.19 |
AC006042.7 |
|
838 |
0.45 |
| chrY_2804797_2805062 | 0.19 |
ZFY |
zinc finger protein, Y-linked |
1383 |
0.54 |
| chr17_7239932_7240900 | 0.19 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
507 |
0.51 |
| chr1_181003153_181003921 | 0.19 |
MR1 |
major histocompatibility complex, class I-related |
398 |
0.84 |
| chr1_205410656_205410807 | 0.19 |
ENSG00000199059 |
. |
6795 |
0.17 |
| chr5_149790034_149790185 | 0.19 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
2186 |
0.28 |
| chr16_11678677_11678828 | 0.18 |
LITAF |
lipopolysaccharide-induced TNF factor |
1477 |
0.42 |
| chr1_24862040_24862255 | 0.18 |
ENSG00000266551 |
. |
5943 |
0.18 |
| chr3_108544304_108544541 | 0.18 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
2803 |
0.36 |
| chr14_22992025_22992229 | 0.18 |
TRAJ15 |
T cell receptor alpha joining 15 |
6453 |
0.11 |
| chr17_56477664_56477815 | 0.18 |
RNF43 |
ring finger protein 43 |
2343 |
0.23 |
| chr3_177077016_177077315 | 0.18 |
ENSG00000252028 |
. |
144175 |
0.05 |
| chr18_13266147_13266338 | 0.18 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
11860 |
0.19 |
| chr22_18250195_18250346 | 0.18 |
ENSG00000264757 |
. |
3245 |
0.23 |
| chr11_128375528_128375679 | 0.18 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
314 |
0.92 |
| chr22_30617296_30617447 | 0.18 |
RP3-438O4.4 |
|
14273 |
0.11 |
| chr6_144472562_144472798 | 0.18 |
STX11 |
syntaxin 11 |
1017 |
0.64 |
| chr11_128387324_128387802 | 0.18 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
4533 |
0.26 |
| chr12_121476370_121477291 | 0.18 |
OASL |
2'-5'-oligoadenylate synthetase-like |
80 |
0.96 |
| chr11_2322055_2322323 | 0.18 |
C11orf21 |
chromosome 11 open reading frame 21 |
954 |
0.34 |
| chr11_95829215_95829366 | 0.18 |
MTMR2 |
myotubularin related protein 2 |
171831 |
0.03 |
| chr18_77283391_77284187 | 0.18 |
AC018445.1 |
Uncharacterized protein |
7732 |
0.29 |
| chr14_78108664_78108852 | 0.17 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
25642 |
0.16 |
| chr9_134608223_134608646 | 0.17 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
4491 |
0.23 |
| chr2_175460831_175461348 | 0.17 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
1404 |
0.41 |
| chr19_1021420_1021571 | 0.17 |
ENSG00000207357 |
. |
26 |
0.83 |
| chr4_154390309_154390666 | 0.17 |
KIAA0922 |
KIAA0922 |
2986 |
0.33 |
| chrX_107070045_107070650 | 0.17 |
MID2 |
midline 2 |
703 |
0.7 |
| chr20_17591518_17591669 | 0.17 |
ENSG00000202260 |
. |
34438 |
0.14 |
| chr1_180469296_180469554 | 0.17 |
ACBD6 |
acyl-CoA binding domain containing 6 |
2664 |
0.28 |
| chr17_5186506_5186842 | 0.17 |
RABEP1 |
rabaptin, RAB GTPase binding effector protein 1 |
892 |
0.56 |
| chr10_13382497_13382864 | 0.17 |
SEPHS1 |
selenophosphate synthetase 1 |
110 |
0.97 |
| chr5_136467995_136468146 | 0.17 |
ENSG00000222285 |
. |
163075 |
0.04 |
| chr14_50053552_50053772 | 0.17 |
RN7SL1 |
RNA, 7SL, cytoplasmic 1 |
364 |
0.55 |
| chr22_40881445_40881596 | 0.17 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
22082 |
0.15 |
| chr19_6738870_6739021 | 0.17 |
TRIP10 |
thyroid hormone receptor interactor 10 |
746 |
0.45 |
| chr2_197040446_197040597 | 0.17 |
STK17B |
serine/threonine kinase 17b |
530 |
0.79 |
| chr19_10381107_10381258 | 0.17 |
ICAM1 |
intercellular adhesion molecule 1 |
329 |
0.72 |
| chr10_82225109_82225470 | 0.17 |
TSPAN14 |
tetraspanin 14 |
6231 |
0.22 |
| chr5_175968739_175969051 | 0.16 |
CDHR2 |
cadherin-related family member 2 |
617 |
0.6 |
| chr13_46742646_46742820 | 0.16 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
79 |
0.95 |
| chr17_74067054_74067205 | 0.16 |
SRP68 |
signal recognition particle 68kDa |
1449 |
0.26 |
| chr19_52832897_52833072 | 0.16 |
ZNF610 |
zinc finger protein 610 |
6514 |
0.11 |
| chr19_33863850_33864075 | 0.16 |
CEBPG |
CCAAT/enhancer binding protein (C/EBP), gamma |
274 |
0.92 |
| chr7_26415346_26415505 | 0.16 |
AC004540.4 |
|
467 |
0.85 |
| chr22_37681278_37681429 | 0.16 |
CYTH4 |
cytohesin 4 |
2825 |
0.23 |
| chr19_42052916_42053110 | 0.16 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
2873 |
0.23 |
| chr2_192070921_192071072 | 0.16 |
MYO1B |
myosin IB |
38915 |
0.17 |
| chr19_10342329_10342480 | 0.16 |
DNMT1 |
DNA (cytosine-5-)-methyltransferase 1 |
442 |
0.39 |
| chr17_25958202_25958709 | 0.16 |
LGALS9 |
lectin, galactoside-binding, soluble, 9 |
222 |
0.92 |
| chr17_43306981_43307138 | 0.16 |
CTD-2020K17.1 |
|
7470 |
0.1 |
| chrX_78343760_78343985 | 0.16 |
GPR174 |
G protein-coupled receptor 174 |
82597 |
0.11 |
| chr2_242810596_242810747 | 0.16 |
CXXC11 |
CXXC finger protein 11 |
1210 |
0.33 |
| chr17_76255043_76255413 | 0.16 |
TMEM235 |
transmembrane protein 235 |
27106 |
0.12 |
| chr5_169723230_169723381 | 0.16 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
1926 |
0.38 |
| chr8_22876543_22876783 | 0.16 |
RP11-875O11.1 |
|
185 |
0.92 |
| chr1_19785278_19785429 | 0.16 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
25226 |
0.14 |
| chr19_7411383_7411820 | 0.16 |
CTB-133G6.1 |
|
2247 |
0.26 |
| chrY_21729398_21729741 | 0.16 |
TXLNG2P |
taxilin gamma 2, pseudogene |
301 |
0.92 |
| chr2_231646247_231646398 | 0.16 |
ENSG00000201044 |
. |
29466 |
0.17 |
| chr5_35863104_35863313 | 0.16 |
IL7R |
interleukin 7 receptor |
6214 |
0.22 |
| chr5_100188986_100189137 | 0.16 |
ENSG00000221263 |
. |
36792 |
0.19 |
| chr12_51782252_51782403 | 0.16 |
GALNT6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
2756 |
0.2 |
| chr5_176728326_176728485 | 0.16 |
RAB24 |
RAB24, member RAS oncogene family |
2302 |
0.17 |
| chr6_12010262_12011396 | 0.15 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
1404 |
0.51 |
| chr6_138776848_138777021 | 0.15 |
ENSG00000266555 |
. |
20503 |
0.19 |
| chr3_16329814_16329965 | 0.15 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
19141 |
0.15 |
| chr16_48642312_48642839 | 0.15 |
N4BP1 |
NEDD4 binding protein 1 |
1545 |
0.4 |
| chr6_47666717_47666868 | 0.15 |
GPR115 |
G protein-coupled receptor 115 |
503 |
0.8 |
| chr12_26999341_26999492 | 0.15 |
ITPR2 |
inositol 1,4,5-trisphosphate receptor, type 2 |
13285 |
0.23 |
| chr7_106505707_106506635 | 0.15 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
247 |
0.95 |
| chr1_899115_899266 | 0.15 |
PLEKHN1 |
pleckstrin homology domain containing, family N member 1 |
2687 |
0.12 |
| chr17_75450826_75450977 | 0.15 |
SEPT9 |
septin 9 |
739 |
0.6 |
| chr1_201323781_201323932 | 0.15 |
TNNT2 |
troponin T type 2 (cardiac) |
18526 |
0.16 |
| chr22_29563318_29563469 | 0.15 |
ENSG00000251952 |
. |
24403 |
0.12 |
| chr2_102972689_102973095 | 0.15 |
IL18R1 |
interleukin 18 receptor 1 |
503 |
0.79 |
| chr6_170403243_170403627 | 0.15 |
RP11-302L19.1 |
|
74306 |
0.11 |
| chr12_92524835_92525431 | 0.15 |
C12orf79 |
chromosome 12 open reading frame 79 |
5664 |
0.19 |
| chr2_68593138_68593303 | 0.15 |
AC015969.3 |
|
504 |
0.63 |
| chrY_15590512_15590663 | 0.15 |
UTY |
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked |
958 |
0.72 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.3 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 0.3 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.3 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.1 | 0.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.3 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.3 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.2 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.2 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.2 | GO:0002517 | T cell tolerance induction(GO:0002517) regulation of T cell tolerance induction(GO:0002664) positive regulation of T cell tolerance induction(GO:0002666) |
| 0.0 | 0.2 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.1 | GO:0002335 | mature B cell differentiation involved in immune response(GO:0002313) mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.4 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.0 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0042088 | T-helper 1 type immune response(GO:0042088) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.2 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0061756 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.1 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.2 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.2 | GO:0000045 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.0 | 0.0 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.0 | GO:0032490 | detection of molecule of bacterial origin(GO:0032490) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.2 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.1 | GO:0070076 | histone lysine demethylation(GO:0070076) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.0 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.7 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.5 | GO:0004629 | phospholipase C activity(GO:0004629) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.0 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.4 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.6 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.0 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.0 | REACTOME FORMATION OF THE HIV1 EARLY ELONGATION COMPLEX | Genes involved in Formation of the HIV-1 Early Elongation Complex |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |