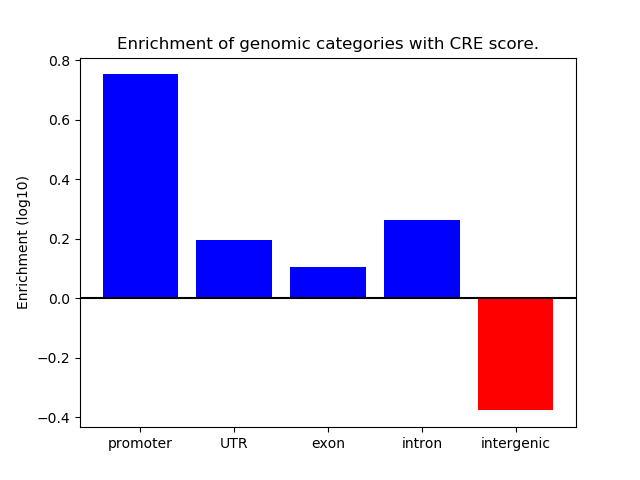
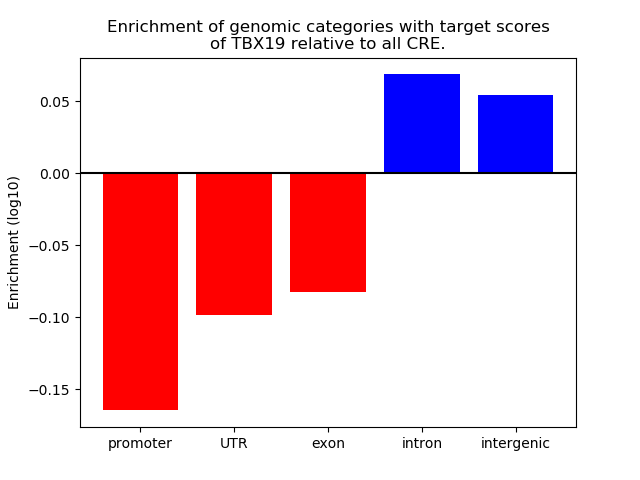
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
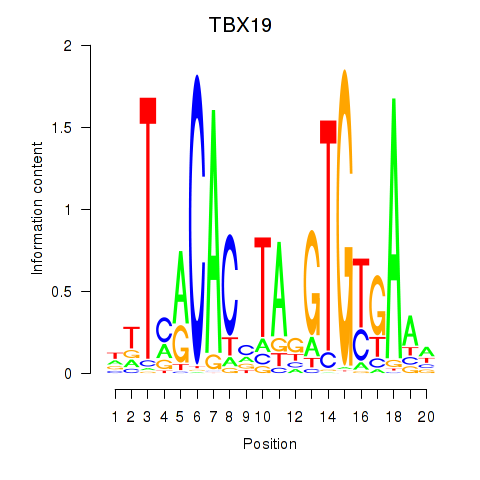
Results for TBX19
Z-value: 0.92
Transcription factors associated with TBX19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX19
|
ENSG00000143178.8 | T-box transcription factor 19 |
Activity of the TBX19 motif across conditions
Conditions sorted by the z-value of the TBX19 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
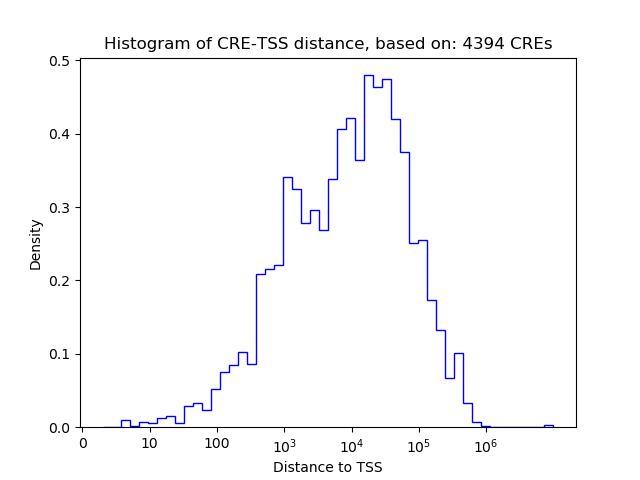
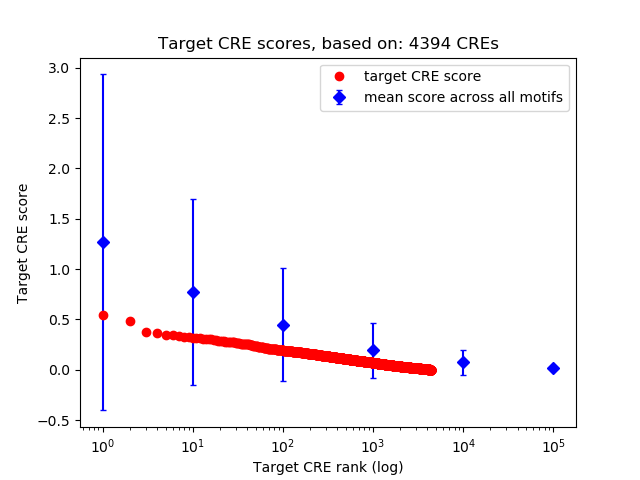
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_85479176_85479377 | 0.54 |
TBX18 |
T-box 18 |
5039 |
0.33 |
| chr8_49832237_49832502 | 0.49 |
SNAI2 |
snail family zinc finger 2 |
1619 |
0.54 |
| chr7_64895070_64895221 | 0.37 |
ZNF92 |
zinc finger protein 92 |
56351 |
0.13 |
| chr8_13371594_13371816 | 0.37 |
DLC1 |
deleted in liver cancer 1 |
569 |
0.79 |
| chr2_151334778_151334948 | 0.35 |
RND3 |
Rho family GTPase 3 |
7033 |
0.34 |
| chr4_81192543_81192855 | 0.34 |
FGF5 |
fibroblast growth factor 5 |
4906 |
0.28 |
| chr7_78398482_78399243 | 0.33 |
MAGI2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
1534 |
0.56 |
| chr18_56558977_56559159 | 0.32 |
ZNF532 |
zinc finger protein 532 |
26282 |
0.16 |
| chr8_89338531_89338740 | 0.32 |
RP11-586K2.1 |
|
430 |
0.76 |
| chr7_19145980_19146451 | 0.32 |
AC003986.6 |
|
5882 |
0.17 |
| chr12_47358106_47358377 | 0.32 |
PCED1B |
PC-esterase domain containing 1B |
115145 |
0.06 |
| chr6_3248086_3248639 | 0.31 |
PSMG4 |
proteasome (prosome, macropain) assembly chaperone 4 |
10766 |
0.18 |
| chr12_3069053_3070004 | 0.31 |
TEAD4 |
TEA domain family member 4 |
445 |
0.77 |
| chr3_98616375_98616538 | 0.31 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
3559 |
0.22 |
| chr2_230035602_230035855 | 0.31 |
PID1 |
phosphotyrosine interaction domain containing 1 |
61073 |
0.14 |
| chr2_9347772_9348251 | 0.30 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
1117 |
0.65 |
| chr19_54057819_54058678 | 0.30 |
ZNF331 |
zinc finger protein 331 |
167 |
0.9 |
| chr13_33858654_33859881 | 0.29 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
625 |
0.76 |
| chr2_144692978_144693129 | 0.29 |
AC016910.1 |
|
1587 |
0.49 |
| chr3_69836526_69836696 | 0.28 |
ENSG00000241665 |
. |
17683 |
0.23 |
| chr3_150132523_150132716 | 0.28 |
TSC22D2 |
TSC22 domain family, member 2 |
3823 |
0.33 |
| chr6_148664293_148664728 | 0.28 |
SASH1 |
SAM and SH3 domain containing 1 |
781 |
0.74 |
| chr10_95239197_95239357 | 0.28 |
MYOF |
myoferlin |
2674 |
0.26 |
| chr7_155592218_155592554 | 0.28 |
SHH |
sonic hedgehog |
9380 |
0.23 |
| chr8_134203052_134203203 | 0.28 |
WISP1 |
WNT1 inducible signaling pathway protein 1 |
155 |
0.97 |
| chr22_20119477_20120671 | 0.28 |
ZDHHC8 |
zinc finger, DHHC-type containing 8 |
603 |
0.53 |
| chr4_188523294_188523560 | 0.28 |
ZFP42 |
ZFP42 zinc finger protein |
393498 |
0.01 |
| chr8_37350646_37351319 | 0.27 |
RP11-150O12.6 |
|
23557 |
0.24 |
| chr4_120970638_120970865 | 0.27 |
RP11-679C8.2 |
|
17362 |
0.21 |
| chr16_64927185_64927365 | 0.27 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
166306 |
0.04 |
| chr3_157262200_157262351 | 0.27 |
C3orf55 |
chromosome 3 open reading frame 55 |
1042 |
0.6 |
| chr5_141931000_141931262 | 0.26 |
ENSG00000252831 |
. |
17221 |
0.21 |
| chr2_8055570_8055940 | 0.26 |
ENSG00000221255 |
. |
338783 |
0.01 |
| chr8_32405226_32405421 | 0.26 |
NRG1 |
neuregulin 1 |
405 |
0.92 |
| chr3_29377379_29377667 | 0.26 |
ENSG00000216169 |
. |
33389 |
0.18 |
| chr10_93391857_93392062 | 0.26 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
852 |
0.74 |
| chr19_15333567_15333831 | 0.26 |
EPHX3 |
epoxide hydrolase 3 |
9544 |
0.16 |
| chr7_25889169_25889395 | 0.25 |
ENSG00000199085 |
. |
100324 |
0.08 |
| chr10_52835110_52835261 | 0.25 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
1251 |
0.52 |
| chr10_60501911_60502062 | 0.25 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
51309 |
0.18 |
| chrX_46186736_46187042 | 0.25 |
KRBOX4 |
KRAB box domain containing 4 |
119403 |
0.06 |
| chr19_16179581_16179939 | 0.25 |
TPM4 |
tropomyosin 4 |
1250 |
0.44 |
| chr7_44144273_44144986 | 0.25 |
AEBP1 |
AE binding protein 1 |
236 |
0.84 |
| chr12_64238850_64239338 | 0.25 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
553 |
0.72 |
| chr8_42063953_42065062 | 0.25 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr2_46928125_46928276 | 0.24 |
SOCS5 |
suppressor of cytokine signaling 5 |
1874 |
0.38 |
| chr5_63933889_63934062 | 0.24 |
FAM159B |
family with sequence similarity 159, member B |
52160 |
0.14 |
| chr20_19956550_19956867 | 0.24 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
41052 |
0.15 |
| chr12_18449684_18450065 | 0.23 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
14896 |
0.31 |
| chr10_95240803_95240954 | 0.23 |
MYOF |
myoferlin |
1073 |
0.51 |
| chr3_147638936_147639087 | 0.23 |
ENSG00000221431 |
. |
9723 |
0.32 |
| chr10_109674932_109675169 | 0.23 |
ENSG00000200079 |
. |
453732 |
0.01 |
| chr10_63991622_63991773 | 0.23 |
RTKN2 |
rhotekin 2 |
4325 |
0.32 |
| chr3_141088214_141088426 | 0.23 |
RP11-438D8.2 |
|
2341 |
0.33 |
| chr2_205411137_205411321 | 0.23 |
PARD3B |
par-3 family cell polarity regulator beta |
506 |
0.89 |
| chr15_60688893_60689153 | 0.23 |
ANXA2 |
annexin A2 |
514 |
0.85 |
| chr22_39801235_39801470 | 0.23 |
TAB1 |
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
5563 |
0.17 |
| chr22_25346267_25346418 | 0.23 |
KIAA1671 |
KIAA1671 |
2355 |
0.29 |
| chr10_13748513_13748974 | 0.22 |
ENSG00000222235 |
. |
1765 |
0.25 |
| chr4_99929189_99929340 | 0.22 |
ENSG00000265213 |
. |
10726 |
0.16 |
| chr3_156873804_156874020 | 0.22 |
ENSG00000201778 |
. |
2575 |
0.21 |
| chr4_187901481_187901677 | 0.22 |
ENSG00000252382 |
. |
122969 |
0.06 |
| chr8_27729804_27730180 | 0.22 |
ENSG00000265847 |
. |
13641 |
0.16 |
| chr3_42094199_42094430 | 0.22 |
TRAK1 |
trafficking protein, kinesin binding 1 |
38248 |
0.19 |
| chr16_69959859_69960167 | 0.22 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
1120 |
0.44 |
| chr19_11784446_11784993 | 0.21 |
CTC-499B15.4 |
|
8642 |
0.14 |
| chr4_170189368_170189538 | 0.21 |
SH3RF1 |
SH3 domain containing ring finger 1 |
1655 |
0.5 |
| chr11_69454351_69454502 | 0.21 |
CCND1 |
cyclin D1 |
1429 |
0.47 |
| chr10_123922760_123923208 | 0.21 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
43 |
0.99 |
| chr6_24584146_24584297 | 0.21 |
KIAA0319 |
KIAA0319 |
61735 |
0.09 |
| chr2_24123591_24123937 | 0.21 |
ATAD2B |
ATPase family, AAA domain containing 2B |
26171 |
0.17 |
| chr3_65582462_65582613 | 0.21 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
873 |
0.76 |
| chr1_61586210_61586441 | 0.21 |
RP4-802A10.1 |
|
4080 |
0.27 |
| chr2_45228184_45228926 | 0.21 |
SIX2 |
SIX homeobox 2 |
8014 |
0.26 |
| chr18_53090404_53090609 | 0.21 |
TCF4 |
transcription factor 4 |
763 |
0.7 |
| chr15_27508765_27508916 | 0.20 |
GABRG3 |
gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
7849 |
0.23 |
| chr2_221940084_221940235 | 0.20 |
EPHA4 |
EPH receptor A4 |
427121 |
0.01 |
| chr2_238319908_238320059 | 0.20 |
COL6A3 |
collagen, type VI, alpha 3 |
2808 |
0.28 |
| chr9_17017802_17018053 | 0.20 |
ENSG00000241152 |
. |
35972 |
0.22 |
| chr3_77090223_77090496 | 0.20 |
ROBO2 |
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
478 |
0.88 |
| chr17_29891051_29891281 | 0.20 |
ENSG00000221038 |
. |
253 |
0.86 |
| chr2_158117199_158117478 | 0.20 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
3228 |
0.3 |
| chr2_39750505_39750656 | 0.20 |
AC007246.3 |
|
4764 |
0.3 |
| chr11_7599067_7599755 | 0.20 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
1140 |
0.49 |
| chr3_168759646_168760009 | 0.20 |
MECOM |
MDS1 and EVI1 complex locus |
85995 |
0.11 |
| chr21_17677767_17678526 | 0.20 |
ENSG00000201025 |
. |
21057 |
0.28 |
| chr11_62306171_62306793 | 0.20 |
RP11-864I4.3 |
|
1698 |
0.19 |
| chr4_158141912_158142089 | 0.20 |
GRIA2 |
glutamate receptor, ionotropic, AMPA 2 |
41 |
0.98 |
| chr2_26981186_26981361 | 0.20 |
SLC35F6 |
solute carrier family 35, member F6 |
5883 |
0.19 |
| chr2_192710921_192711254 | 0.20 |
AC098617.1 |
|
178 |
0.9 |
| chr4_189471544_189471830 | 0.20 |
ENSG00000252275 |
. |
166192 |
0.04 |
| chr9_19378948_19379099 | 0.20 |
RPS6 |
ribosomal protein S6 |
1173 |
0.37 |
| chr7_73443076_73443679 | 0.20 |
ELN |
elastin |
859 |
0.62 |
| chr3_46854432_46854583 | 0.20 |
PRSS50 |
protease, serine, 50 |
443 |
0.45 |
| chr4_46935635_46935786 | 0.20 |
COX7B2 |
cytochrome c oxidase subunit VIIb2 |
24458 |
0.22 |
| chr13_42422903_42423054 | 0.20 |
ENSG00000241406 |
. |
41165 |
0.17 |
| chr17_2499771_2499922 | 0.19 |
PAFAH1B1 |
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa) |
2806 |
0.23 |
| chr8_126557270_126557537 | 0.19 |
ENSG00000266452 |
. |
100596 |
0.08 |
| chr18_57170103_57170344 | 0.19 |
CCBE1 |
collagen and calcium binding EGF domains 1 |
22751 |
0.19 |
| chr2_119989088_119989281 | 0.19 |
STEAP3 |
STEAP family member 3, metalloreductase |
7755 |
0.2 |
| chr8_93090595_93090746 | 0.19 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
2306 |
0.45 |
| chr6_69346067_69346218 | 0.19 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
883 |
0.52 |
| chr2_190017431_190017582 | 0.19 |
ENSG00000264725 |
. |
19669 |
0.19 |
| chr3_42922021_42922670 | 0.19 |
CYP8B1 |
cytochrome P450, family 8, subfamily B, polypeptide 1 |
4712 |
0.15 |
| chr5_110846365_110846666 | 0.19 |
STARD4-AS1 |
STARD4 antisense RNA 1 |
1409 |
0.37 |
| chr2_26240891_26241042 | 0.19 |
AC013449.1 |
Uncharacterized protein |
10515 |
0.17 |
| chr2_33361191_33361342 | 0.19 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
1542 |
0.54 |
| chr15_59560315_59560475 | 0.19 |
RP11-429D19.1 |
|
2966 |
0.2 |
| chr1_51069078_51069376 | 0.19 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
8943 |
0.23 |
| chr5_159510080_159510300 | 0.19 |
PWWP2A |
PWWP domain containing 2A |
36239 |
0.14 |
| chr13_100635852_100636003 | 0.19 |
ZIC2 |
Zic family member 2 |
1901 |
0.33 |
| chr3_148734494_148734920 | 0.19 |
GYG1 |
glycogenin 1 |
24549 |
0.16 |
| chr4_110901886_110902148 | 0.19 |
ENSG00000207260 |
. |
11464 |
0.24 |
| chr8_125212210_125212404 | 0.19 |
FER1L6-AS2 |
FER1L6 antisense RNA 2 |
28544 |
0.2 |
| chr7_55322762_55323341 | 0.19 |
EGFR-AS1 |
EGFR antisense RNA 1 |
66424 |
0.13 |
| chr16_81037098_81037249 | 0.19 |
CENPN |
centromere protein N |
2930 |
0.16 |
| chr3_25193145_25193356 | 0.19 |
RARB |
retinoic acid receptor, beta |
22573 |
0.26 |
| chr13_51971753_51971904 | 0.19 |
INTS6 |
integrator complex subunit 6 |
12428 |
0.15 |
| chr3_170136599_170137401 | 0.18 |
CLDN11 |
claudin 11 |
145 |
0.97 |
| chr19_47735595_47736220 | 0.18 |
BBC3 |
BCL2 binding component 3 |
116 |
0.95 |
| chr18_53067346_53067650 | 0.18 |
TCF4 |
transcription factor 4 |
1068 |
0.61 |
| chr5_111090218_111090459 | 0.18 |
NREP |
neuronal regeneration related protein |
1610 |
0.42 |
| chr16_3163066_3163293 | 0.18 |
ZNF205 |
zinc finger protein 205 |
383 |
0.54 |
| chr4_63575291_63575489 | 0.18 |
ENSG00000265085 |
. |
114295 |
0.07 |
| chr2_36600561_36600712 | 0.18 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
17022 |
0.28 |
| chr6_18551314_18551465 | 0.18 |
ENSG00000207775 |
. |
20626 |
0.27 |
| chr2_39964531_39964918 | 0.18 |
THUMPD2 |
THUMP domain containing 2 |
41565 |
0.16 |
| chr3_55520023_55520469 | 0.18 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1085 |
0.48 |
| chr22_34249489_34249782 | 0.18 |
LARGE |
like-glycosyltransferase |
7977 |
0.27 |
| chr12_50415148_50415300 | 0.18 |
RACGAP1 |
Rac GTPase activating protein 1 |
3959 |
0.15 |
| chr7_55196777_55196928 | 0.18 |
EGFR |
epidermal growth factor receptor |
19312 |
0.25 |
| chr22_31218163_31218918 | 0.18 |
OSBP2 |
oxysterol binding protein 2 |
37 |
0.98 |
| chr14_62169704_62169855 | 0.18 |
HIF1A |
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) |
5439 |
0.26 |
| chr3_111487167_111487318 | 0.18 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
35898 |
0.18 |
| chr5_82864302_82864453 | 0.18 |
VCAN-AS1 |
VCAN antisense RNA 1 |
6244 |
0.28 |
| chr8_57594130_57594603 | 0.18 |
RP11-17A4.2 |
|
192709 |
0.03 |
| chr5_72630160_72630334 | 0.18 |
FOXD1 |
forkhead box D1 |
114105 |
0.06 |
| chr3_61833858_61834009 | 0.18 |
ENSG00000252420 |
. |
109030 |
0.08 |
| chr5_55193328_55193500 | 0.18 |
AC008914.1 |
Uncharacterized protein |
31155 |
0.14 |
| chr13_75346514_75346665 | 0.18 |
ENSG00000206812 |
. |
337135 |
0.01 |
| chr15_68573463_68573639 | 0.18 |
FEM1B |
fem-1 homolog b (C. elegans) |
1254 |
0.47 |
| chrX_139173235_139173445 | 0.18 |
ENSG00000241248 |
. |
5326 |
0.31 |
| chr1_114522595_114522893 | 0.18 |
OLFML3 |
olfactomedin-like 3 |
49 |
0.97 |
| chr2_165463349_165463500 | 0.18 |
GRB14 |
growth factor receptor-bound protein 14 |
13043 |
0.27 |
| chr12_50608832_50609096 | 0.18 |
RP3-405J10.4 |
|
4423 |
0.14 |
| chr6_111925125_111925433 | 0.18 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
1795 |
0.32 |
| chr10_121418043_121418219 | 0.18 |
BAG3 |
BCL2-associated athanogene 3 |
2009 |
0.38 |
| chr6_18434642_18434820 | 0.18 |
ENSG00000238458 |
. |
32448 |
0.19 |
| chr2_42866321_42866472 | 0.18 |
ENSG00000194270 |
. |
1295 |
0.53 |
| chr4_125866592_125866796 | 0.17 |
ANKRD50 |
ankyrin repeat domain 50 |
232807 |
0.02 |
| chr4_54172326_54172477 | 0.17 |
ENSG00000207385 |
. |
40667 |
0.17 |
| chr1_17871786_17871970 | 0.17 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
5548 |
0.27 |
| chr9_101706823_101707024 | 0.17 |
RP11-92C4.6 |
|
866 |
0.45 |
| chr10_33425320_33425471 | 0.17 |
ENSG00000263576 |
. |
37831 |
0.16 |
| chr2_203881741_203882170 | 0.17 |
NBEAL1 |
neurobeachin-like 1 |
2353 |
0.35 |
| chr2_101358636_101358796 | 0.17 |
NPAS2 |
neuronal PAS domain protein 2 |
77898 |
0.09 |
| chr4_26347299_26347489 | 0.17 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
2632 |
0.42 |
| chr3_73652541_73652692 | 0.17 |
ENSG00000239119 |
. |
9463 |
0.21 |
| chr13_52132399_52132573 | 0.17 |
ENSG00000266611 |
. |
5761 |
0.14 |
| chr1_173835540_173835691 | 0.17 |
ENSG00000234741 |
. |
106 |
0.88 |
| chr9_90812890_90813286 | 0.17 |
ENSG00000252299 |
. |
176096 |
0.03 |
| chr4_122616322_122616745 | 0.17 |
ANXA5 |
annexin A5 |
1584 |
0.45 |
| chr8_96714886_96715046 | 0.17 |
ENSG00000223297 |
. |
23823 |
0.26 |
| chr2_190041307_190041749 | 0.17 |
COL5A2 |
collagen, type V, alpha 2 |
3077 |
0.31 |
| chr1_51726454_51726959 | 0.17 |
RP11-296A18.6 |
|
5040 |
0.16 |
| chr1_55865474_55865625 | 0.17 |
ENSG00000252162 |
. |
1362 |
0.5 |
| chr13_50645613_50645841 | 0.17 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
10580 |
0.16 |
| chr12_91487813_91488090 | 0.17 |
LUM |
lumican |
17657 |
0.21 |
| chr9_99265347_99265498 | 0.17 |
HABP4 |
hyaluronan binding protein 4 |
52935 |
0.13 |
| chr6_157270459_157270621 | 0.17 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
48033 |
0.19 |
| chr2_142888867_142889088 | 0.17 |
LRP1B |
low density lipoprotein receptor-related protein 1B |
293 |
0.93 |
| chr7_72933105_72933537 | 0.17 |
BAZ1B |
bromodomain adjacent to zinc finger domain, 1B |
3281 |
0.19 |
| chr10_34630198_34630349 | 0.17 |
PARD3 |
par-3 family cell polarity regulator |
31188 |
0.25 |
| chr13_33778212_33778569 | 0.17 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
1753 |
0.42 |
| chr9_124498552_124498751 | 0.17 |
DAB2IP |
DAB2 interacting protein |
6413 |
0.28 |
| chr14_100345979_100346191 | 0.17 |
EML1 |
echinoderm microtubule associated protein like 1 |
14139 |
0.22 |
| chr2_64633040_64633281 | 0.17 |
AC008074.3 |
|
47772 |
0.13 |
| chr4_142423380_142423531 | 0.17 |
IL15 |
interleukin 15 |
134297 |
0.05 |
| chr7_130623665_130623952 | 0.16 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
45060 |
0.16 |
| chr20_32149890_32150174 | 0.16 |
CBFA2T2 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 2 |
108 |
0.97 |
| chr7_94027821_94027972 | 0.16 |
COL1A2 |
collagen, type I, alpha 2 |
4023 |
0.33 |
| chr15_96855709_96855975 | 0.16 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
10032 |
0.16 |
| chr15_37179413_37179956 | 0.16 |
ENSG00000212511 |
. |
34843 |
0.22 |
| chr17_36600608_36600759 | 0.16 |
ENSG00000260833 |
. |
7249 |
0.16 |
| chr4_87518747_87518898 | 0.16 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
1976 |
0.35 |
| chr3_149266255_149266548 | 0.16 |
ENSG00000252581 |
. |
22983 |
0.17 |
| chr17_57505253_57505417 | 0.16 |
RP11-567L7.5 |
|
24671 |
0.18 |
| chrX_37730532_37730793 | 0.16 |
ENSG00000206983 |
. |
1300 |
0.5 |
| chr1_198791306_198791457 | 0.16 |
ENSG00000207975 |
. |
36730 |
0.18 |
| chr17_53809217_53809377 | 0.16 |
TMEM100 |
transmembrane protein 100 |
185 |
0.96 |
| chr1_172320901_172321057 | 0.16 |
ENSG00000252354 |
. |
3696 |
0.22 |
| chr9_16871555_16871924 | 0.16 |
BNC2 |
basonuclin 2 |
898 |
0.74 |
| chr22_33198587_33198826 | 0.16 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
1019 |
0.61 |
| chr8_25901085_25901478 | 0.16 |
EBF2 |
early B-cell factor 2 |
1632 |
0.53 |
| chr16_50241915_50242069 | 0.16 |
ADCY7 |
adenylate cyclase 7 |
38056 |
0.13 |
| chr1_87583813_87583964 | 0.16 |
ENSG00000221222 |
. |
33093 |
0.21 |
| chr1_202509205_202509449 | 0.16 |
ENSG00000253042 |
. |
12772 |
0.21 |
| chr15_66795614_66795765 | 0.16 |
ENSG00000200623 |
. |
37 |
0.82 |
| chr14_45559122_45559273 | 0.16 |
ENSG00000212615 |
. |
1751 |
0.24 |
| chr11_65202266_65202417 | 0.16 |
ENSG00000245532 |
. |
9588 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.3 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.1 | 0.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 | 0.2 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0090037 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0072283 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.0 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:1903020 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.0 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.0 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:1901862 | negative regulation of striated muscle tissue development(GO:0045843) negative regulation of muscle tissue development(GO:1901862) |
| 0.0 | 0.0 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.2 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 1.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0030553 | cGMP binding(GO:0030553) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY TGF BETA RECEPTOR COMPLEX | Genes involved in Signaling by TGF-beta Receptor Complex |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.0 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |