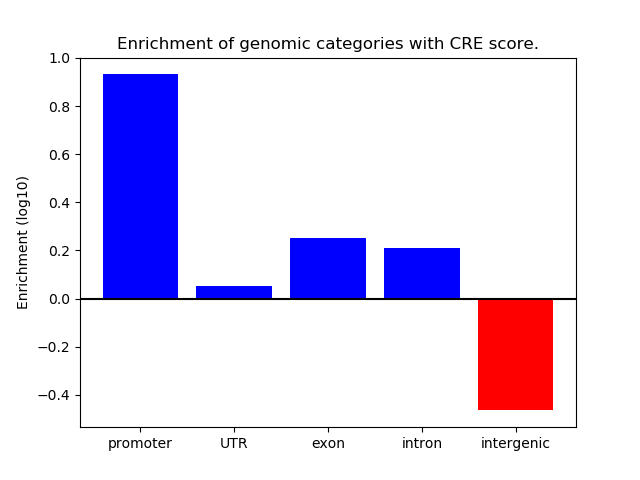
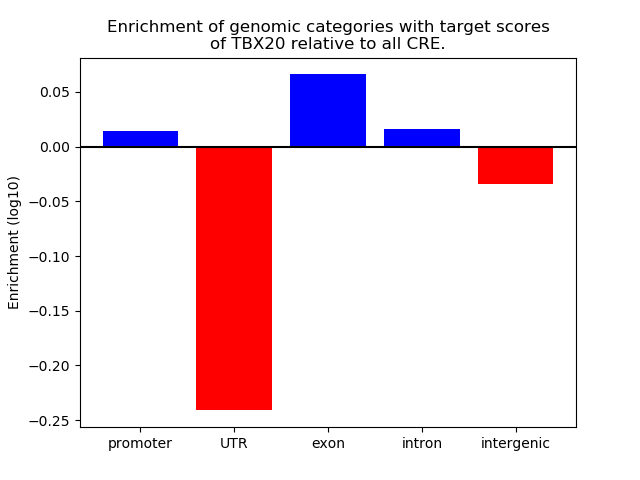
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for TBX20
Z-value: 0.53
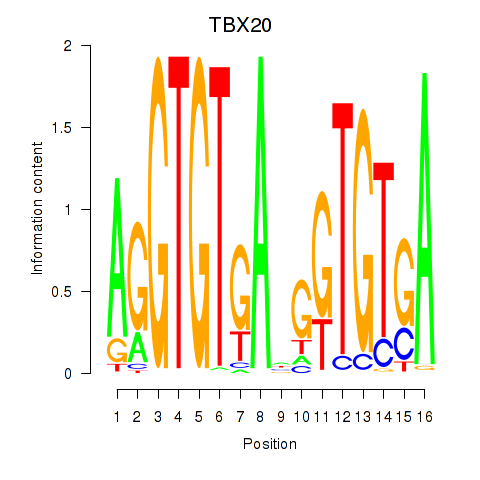
Transcription factors associated with TBX20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX20
|
ENSG00000164532.10 | T-box transcription factor 20 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_35460479_35460660 | TBX20 | 166811 | 0.035710 | -0.80 | 1.0e-02 | Click! |
| chr7_35460237_35460417 | TBX20 | 166569 | 0.035803 | -0.27 | 4.8e-01 | Click! |
| chr7_35293220_35293371 | TBX20 | 463 | 0.886829 | -0.25 | 5.2e-01 | Click! |
| chr7_35293388_35293792 | TBX20 | 168 | 0.972359 | 0.17 | 6.6e-01 | Click! |
| chr7_35293884_35294083 | TBX20 | 225 | 0.960862 | 0.10 | 7.9e-01 | Click! |
Activity of the TBX20 motif across conditions
Conditions sorted by the z-value of the TBX20 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_49626686_49627168 | 0.33 |
TUBA1C |
tubulin, alpha 1c |
5218 |
0.14 |
| chrX_1388495_1388646 | 0.33 |
CSF2RA |
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) |
832 |
0.46 |
| chrY_1338495_1338646 | 0.33 |
NA |
NA |
> 106 |
NA |
| chr4_166177201_166177361 | 0.25 |
ENSG00000200974 |
. |
3549 |
0.24 |
| chr1_14924992_14926197 | 0.24 |
KAZN |
kazrin, periplakin interacting protein |
381 |
0.93 |
| chr18_33389268_33389419 | 0.23 |
ENSG00000207797 |
. |
95546 |
0.08 |
| chrX_67913478_67914353 | 0.23 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
422 |
0.91 |
| chr8_23396205_23396870 | 0.21 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
9964 |
0.16 |
| chr1_20927846_20928118 | 0.21 |
CDA |
cytidine deaminase |
12541 |
0.16 |
| chr11_77160479_77160755 | 0.21 |
DKFZP434E1119 |
|
23799 |
0.18 |
| chr11_1339768_1339919 | 0.21 |
TOLLIP-AS1 |
TOLLIP antisense RNA 1 (head to head) |
8844 |
0.15 |
| chr3_151036628_151036780 | 0.20 |
GPR87 |
G protein-coupled receptor 87 |
1964 |
0.3 |
| chr11_47397838_47397989 | 0.20 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
2029 |
0.17 |
| chr14_24041282_24041433 | 0.20 |
JPH4 |
junctophilin 4 |
853 |
0.43 |
| chr17_74323765_74323916 | 0.20 |
ENSG00000238418 |
. |
3563 |
0.13 |
| chr21_40106063_40106708 | 0.20 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
70846 |
0.12 |
| chr1_185346204_185346355 | 0.20 |
ENSG00000252407 |
. |
57097 |
0.12 |
| chr11_15643095_15643246 | 0.19 |
ENSG00000253072 |
. |
139731 |
0.05 |
| chr13_72371766_72371917 | 0.19 |
DACH1 |
dachshund homolog 1 (Drosophila) |
69066 |
0.14 |
| chr2_176971216_176971605 | 0.19 |
HOXD11 |
homeobox D11 |
604 |
0.48 |
| chrX_65238714_65238865 | 0.18 |
ENSG00000207939 |
. |
77 |
0.98 |
| chr4_54784006_54784157 | 0.18 |
RP11-89B16.1 |
|
18043 |
0.26 |
| chr10_64380282_64380433 | 0.18 |
ZNF365 |
zinc finger protein 365 |
23176 |
0.22 |
| chr19_18485571_18485722 | 0.17 |
GDF15 |
growth differentiation factor 15 |
105 |
0.93 |
| chr6_111180837_111181007 | 0.17 |
ENSG00000199360 |
. |
3302 |
0.2 |
| chr22_36831926_36832077 | 0.17 |
ENSG00000252575 |
. |
2970 |
0.21 |
| chr2_47284763_47284914 | 0.17 |
AC073283.7 |
|
10123 |
0.19 |
| chr12_96801988_96802139 | 0.17 |
CDK17 |
cyclin-dependent kinase 17 |
7725 |
0.2 |
| chr19_54641519_54642181 | 0.17 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
401 |
0.66 |
| chr3_38143707_38143858 | 0.17 |
ENSG00000201965 |
. |
23103 |
0.13 |
| chr7_139467090_139467241 | 0.16 |
HIPK2 |
homeodomain interacting protein kinase 2 |
10352 |
0.23 |
| chr5_139971225_139971376 | 0.16 |
ENSG00000200235 |
. |
12048 |
0.08 |
| chr2_46072609_46072760 | 0.16 |
PRKCE |
protein kinase C, epsilon |
155357 |
0.04 |
| chrX_18686624_18686775 | 0.16 |
RS1 |
retinoschisin 1 |
3530 |
0.25 |
| chr6_12242031_12242182 | 0.16 |
EDN1 |
endothelin 1 |
48490 |
0.18 |
| chr5_126148111_126148262 | 0.15 |
LMNB1 |
lamin B1 |
35305 |
0.19 |
| chr18_53254621_53254995 | 0.15 |
TCF4 |
transcription factor 4 |
461 |
0.89 |
| chr17_29036450_29037561 | 0.15 |
ENSG00000241631 |
. |
7212 |
0.17 |
| chr11_1802236_1802476 | 0.15 |
CTSD |
cathepsin D |
17134 |
0.08 |
| chr13_46507059_46507210 | 0.15 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
36671 |
0.18 |
| chr6_15288185_15288336 | 0.15 |
ENSG00000201367 |
. |
26891 |
0.16 |
| chr8_122650853_122651057 | 0.14 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
578 |
0.78 |
| chr13_84456825_84456976 | 0.14 |
SLITRK1 |
SLIT and NTRK-like family, member 1 |
372 |
0.93 |
| chr11_19391979_19392130 | 0.14 |
NAV2-IT1 |
NAV2 intronic transcript 1 (non-protein coding) |
9977 |
0.19 |
| chr15_52586840_52586991 | 0.14 |
MYO5C |
myosin VC |
1051 |
0.54 |
| chr11_15136549_15136719 | 0.14 |
INSC |
inscuteable homolog (Drosophila) |
150 |
0.97 |
| chr1_235054337_235054488 | 0.14 |
ENSG00000239690 |
. |
14479 |
0.26 |
| chr15_45933944_45934448 | 0.13 |
SQRDL |
sulfide quinone reductase-like (yeast) |
3883 |
0.22 |
| chrX_18443769_18444588 | 0.13 |
CDKL5 |
cyclin-dependent kinase-like 5 |
475 |
0.85 |
| chr4_40630958_40631836 | 0.13 |
RBM47 |
RNA binding motif protein 47 |
484 |
0.87 |
| chr5_179495571_179495863 | 0.13 |
RNF130 |
ring finger protein 130 |
3004 |
0.28 |
| chr19_59086860_59087586 | 0.13 |
MZF1 |
myeloid zinc finger 1 |
2281 |
0.14 |
| chr2_69390525_69390686 | 0.13 |
ENSG00000251850 |
. |
18530 |
0.18 |
| chr7_73686672_73686869 | 0.13 |
ENSG00000252538 |
. |
13824 |
0.15 |
| chr12_4699931_4700082 | 0.13 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
444 |
0.81 |
| chr17_26500569_26500720 | 0.13 |
PYY2 |
peptide YY, 2 (pseudogene) |
53688 |
0.09 |
| chr6_133034939_133035106 | 0.13 |
VNN1 |
vanin 1 |
166 |
0.94 |
| chr17_32575509_32575660 | 0.13 |
CCL2 |
chemokine (C-C motif) ligand 2 |
6720 |
0.14 |
| chr9_123638194_123638739 | 0.13 |
PHF19 |
PHD finger protein 19 |
245 |
0.92 |
| chr11_47397562_47397713 | 0.12 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
2305 |
0.16 |
| chr7_143083179_143083339 | 0.12 |
ZYX |
zyxin |
3166 |
0.14 |
| chr11_33913285_33913515 | 0.12 |
LMO2 |
LIM domain only 2 (rhombotin-like 1) |
436 |
0.85 |
| chr16_19130112_19130751 | 0.12 |
RP11-626G11.3 |
|
2520 |
0.23 |
| chr5_171301439_171301590 | 0.12 |
SMIM23 |
small integral membrane protein 23 |
88624 |
0.08 |
| chr2_66668625_66669005 | 0.12 |
AC092669.1 |
|
153 |
0.89 |
| chr5_147284118_147284269 | 0.12 |
C5orf46 |
chromosome 5 open reading frame 46 |
1872 |
0.29 |
| chr6_45303403_45303554 | 0.12 |
RUNX2 |
runt-related transcription factor 2 |
7014 |
0.24 |
| chr2_8442112_8442263 | 0.12 |
AC011747.7 |
|
373709 |
0.01 |
| chr15_101779077_101779228 | 0.11 |
CHSY1 |
chondroitin sulfate synthase 1 |
12985 |
0.18 |
| chr1_27882831_27883103 | 0.11 |
RP1-159A19.4 |
|
30651 |
0.13 |
| chr14_81888288_81888439 | 0.11 |
STON2 |
stonin 2 |
5385 |
0.33 |
| chr3_174159101_174159924 | 0.11 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
735 |
0.81 |
| chr3_49370394_49370545 | 0.11 |
USP4 |
ubiquitin specific peptidase 4 (proto-oncogene) |
6997 |
0.12 |
| chrX_15933872_15934023 | 0.11 |
ENSG00000200566 |
. |
479 |
0.86 |
| chr10_98424597_98424822 | 0.11 |
PIK3AP1 |
phosphoinositide-3-kinase adaptor protein 1 |
4659 |
0.24 |
| chr3_114477625_114477831 | 0.11 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
59 |
0.98 |
| chr21_36420108_36421159 | 0.11 |
RUNX1 |
runt-related transcription factor 1 |
829 |
0.77 |
| chr6_109702291_109703187 | 0.11 |
CD164 |
CD164 molecule, sialomucin |
164 |
0.91 |
| chr5_40487521_40487672 | 0.11 |
ENSG00000265615 |
. |
167176 |
0.03 |
| chr15_101462786_101462937 | 0.11 |
LRRK1 |
leucine-rich repeat kinase 1 |
3311 |
0.24 |
| chr22_30575363_30575514 | 0.10 |
RP3-438O4.4 |
|
27660 |
0.13 |
| chr5_133860075_133861084 | 0.10 |
JADE2 |
jade family PHD finger 2 |
576 |
0.71 |
| chr12_124252082_124252425 | 0.10 |
DNAH10 |
dynein, axonemal, heavy chain 10 |
5211 |
0.19 |
| chr1_245215310_245215673 | 0.10 |
ENSG00000251754 |
. |
8261 |
0.17 |
| chr15_67334428_67334662 | 0.10 |
SMAD3 |
SMAD family member 3 |
21556 |
0.25 |
| chr2_235789593_235789754 | 0.10 |
SH3BP4 |
SH3-domain binding protein 4 |
70944 |
0.14 |
| chr16_12894626_12894777 | 0.10 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
3007 |
0.25 |
| chr2_175546375_175546968 | 0.10 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
928 |
0.66 |
| chr19_14315927_14317134 | 0.10 |
LPHN1 |
latrophilin 1 |
451 |
0.77 |
| chr19_54813162_54813313 | 0.10 |
LILRA3 |
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
3285 |
0.12 |
| chr10_75626788_75626939 | 0.10 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
6251 |
0.12 |
| chr5_36460091_36460242 | 0.09 |
ENSG00000222178 |
. |
25279 |
0.24 |
| chr5_6632169_6633365 | 0.09 |
NSUN2 |
NOP2/Sun RNA methyltransferase family, member 2 |
388 |
0.68 |
| chr15_85258526_85259319 | 0.09 |
SEC11A |
SEC11 homolog A (S. cerevisiae) |
372 |
0.81 |
| chr1_44443266_44443477 | 0.09 |
B4GALT2 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
1495 |
0.22 |
| chrX_41308838_41309328 | 0.09 |
NYX |
nyctalopin |
2370 |
0.34 |
| chr19_57276717_57276915 | 0.09 |
AC006115.3 |
|
75 |
0.97 |
| chr17_38480138_38480579 | 0.09 |
RARA |
retinoic acid receptor, alpha |
5821 |
0.12 |
| chr19_58769898_58770049 | 0.09 |
CTD-3138B18.6 |
|
8028 |
0.1 |
| chr10_111834916_111835067 | 0.09 |
ADD3 |
adducin 3 (gamma) |
67269 |
0.1 |
| chrX_106045541_106045714 | 0.09 |
TBC1D8B |
TBC1 domain family, member 8B (with GRAM domain) |
283 |
0.94 |
| chr3_53200486_53200762 | 0.09 |
PRKCD |
protein kinase C, delta |
1479 |
0.38 |
| chr20_49154846_49154997 | 0.09 |
ENSG00000239742 |
. |
19614 |
0.13 |
| chr14_21249278_21249626 | 0.09 |
RNASE6 |
ribonuclease, RNase A family, k6 |
242 |
0.86 |
| chr5_14166101_14166252 | 0.09 |
TRIO |
trio Rho guanine nucleotide exchange factor |
17731 |
0.3 |
| chr10_101702164_101702315 | 0.09 |
DNMBP |
dynamin binding protein |
11486 |
0.19 |
| chr1_232715037_232715188 | 0.09 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
17808 |
0.28 |
| chr19_45147190_45148055 | 0.09 |
PVR |
poliovirus receptor |
225 |
0.9 |
| chr6_15422733_15422884 | 0.09 |
JARID2 |
jumonji, AT rich interactive domain 2 |
21719 |
0.24 |
| chr2_231622260_231622411 | 0.09 |
ENSG00000201044 |
. |
5479 |
0.24 |
| chr20_48393006_48393208 | 0.09 |
ENSG00000252540 |
. |
18241 |
0.16 |
| chr17_38615265_38615416 | 0.09 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
15627 |
0.13 |
| chr8_28974548_28974699 | 0.09 |
CTD-2647L4.1 |
|
6533 |
0.16 |
| chr9_42490542_42490693 | 0.08 |
ENSG00000266353 |
. |
89356 |
0.08 |
| chr4_154003849_154004000 | 0.08 |
TRIM2 |
tripartite motif containing 2 |
69721 |
0.12 |
| chr9_126595619_126595770 | 0.08 |
DENND1A |
DENN/MADD domain containing 1A |
96692 |
0.07 |
| chr7_31923226_31923699 | 0.08 |
ENSG00000223070 |
. |
89600 |
0.1 |
| chr8_12909327_12909478 | 0.08 |
ENSG00000206996 |
. |
7543 |
0.26 |
| chr9_129296069_129296220 | 0.08 |
ENSG00000221768 |
. |
2874 |
0.28 |
| chr7_134540880_134541057 | 0.08 |
CALD1 |
caldesmon 1 |
10624 |
0.29 |
| chr1_235098778_235099079 | 0.08 |
ENSG00000239690 |
. |
58995 |
0.14 |
| chr3_8152824_8153002 | 0.08 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
94919 |
0.09 |
| chr15_67351342_67351493 | 0.08 |
SMAD3 |
SMAD family member 3 |
4684 |
0.32 |
| chr11_100719606_100719757 | 0.08 |
ENSG00000200047 |
. |
9320 |
0.23 |
| chr2_240971693_240971844 | 0.08 |
OR6B2 |
olfactory receptor, family 6, subfamily B, member 2 |
1862 |
0.27 |
| chr2_99069835_99070173 | 0.08 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
8591 |
0.24 |
| chr2_219254408_219254559 | 0.08 |
SLC11A1 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
7451 |
0.09 |
| chr9_110624590_110624741 | 0.08 |
ENSG00000244104 |
. |
50470 |
0.13 |
| chr5_83671503_83671654 | 0.08 |
EDIL3 |
EGF-like repeats and discoidin I-like domains 3 |
8626 |
0.25 |
| chr8_126448005_126448156 | 0.08 |
TRIB1 |
tribbles pseudokinase 1 |
2292 |
0.34 |
| chr2_179669166_179669317 | 0.08 |
TTN |
titin |
2907 |
0.23 |
| chr1_65886473_65887602 | 0.08 |
LEPR |
leptin receptor |
634 |
0.48 |
| chr10_75758743_75758953 | 0.08 |
VCL |
vinculin |
974 |
0.53 |
| chr18_60917027_60917178 | 0.08 |
ENSG00000238988 |
. |
55204 |
0.11 |
| chr11_128589269_128589524 | 0.08 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23478 |
0.17 |
| chr10_111978401_111978552 | 0.08 |
MXI1 |
MAX interactor 1, dimerization protein |
7275 |
0.22 |
| chr21_39658446_39658688 | 0.08 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
9912 |
0.28 |
| chr19_41833878_41834134 | 0.08 |
CCDC97 |
coiled-coil domain containing 97 |
8368 |
0.1 |
| chr7_35839522_35840298 | 0.08 |
SEPT7 |
septin 7 |
632 |
0.78 |
| chr3_187662632_187662783 | 0.07 |
BCL6 |
B-cell CLL/lymphoma 6 |
199192 |
0.02 |
| chr3_141205962_141207110 | 0.07 |
RASA2 |
RAS p21 protein activator 2 |
645 |
0.76 |
| chr10_45870649_45870800 | 0.07 |
ALOX5 |
arachidonate 5-lipoxygenase |
1049 |
0.61 |
| chr16_55090478_55090815 | 0.07 |
IRX5 |
iroquois homeobox 5 |
124662 |
0.06 |
| chr13_50697652_50698307 | 0.07 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
41672 |
0.15 |
| chr10_104946109_104946260 | 0.07 |
NT5C2 |
5'-nucleotidase, cytosolic II |
6871 |
0.23 |
| chr4_15907365_15907907 | 0.07 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
32335 |
0.17 |
| chr6_83552799_83553085 | 0.07 |
RP11-445L13__B.3 |
|
89378 |
0.1 |
| chr3_150996526_150996677 | 0.07 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
346 |
0.87 |
| chr1_234857540_234857879 | 0.07 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
112438 |
0.06 |
| chr4_84139267_84139418 | 0.07 |
COQ2 |
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
66576 |
0.11 |
| chr14_23367490_23367641 | 0.07 |
RBM23 |
RNA binding motif protein 23 |
3885 |
0.09 |
| chr12_14796282_14796433 | 0.07 |
RP11-174G6.1 |
|
22232 |
0.16 |
| chr3_38310284_38310435 | 0.07 |
SLC22A13 |
solute carrier family 22 (organic anion/urate transporter), member 13 |
2962 |
0.23 |
| chr3_171899758_171899984 | 0.07 |
FNDC3B |
fibronectin type III domain containing 3B |
55107 |
0.13 |
| chr19_39889302_39889453 | 0.07 |
MED29 |
mediator complex subunit 29 |
7325 |
0.08 |
| chr17_73098532_73098683 | 0.07 |
ARMC7 |
armadillo repeat containing 7 |
7440 |
0.09 |
| chr12_46785265_46785416 | 0.07 |
SLC38A2 |
solute carrier family 38, member 2 |
18690 |
0.22 |
| chr2_75077032_75077263 | 0.07 |
HK2 |
hexokinase 2 |
14850 |
0.24 |
| chr7_37711546_37711697 | 0.07 |
GPR141 |
G protein-coupled receptor 141 |
11779 |
0.23 |
| chr1_183164302_183164453 | 0.07 |
LAMC2 |
laminin, gamma 2 |
8954 |
0.24 |
| chr5_124084650_124085032 | 0.07 |
ZNF608 |
zinc finger protein 608 |
341 |
0.83 |
| chr13_47250015_47250166 | 0.07 |
LRCH1 |
leucine-rich repeats and calponin homology (CH) domain containing 1 |
3881 |
0.34 |
| chr18_22864008_22864159 | 0.07 |
CTD-2006O16.2 |
|
18007 |
0.26 |
| chr19_17345728_17346858 | 0.07 |
OCEL1 |
occludin/ELL domain containing 1 |
7228 |
0.08 |
| chr12_69742009_69742310 | 0.06 |
LYZ |
lysozyme |
5 |
0.97 |
| chr5_95205846_95206289 | 0.06 |
ENSG00000250362 |
. |
8577 |
0.15 |
| chr1_33811326_33811735 | 0.06 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
3956 |
0.14 |
| chr1_203256231_203256411 | 0.06 |
BTG2 |
BTG family, member 2 |
18343 |
0.16 |
| chr3_172230855_172231028 | 0.06 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
10324 |
0.25 |
| chr17_32573039_32573288 | 0.06 |
CCL2 |
chemokine (C-C motif) ligand 2 |
9141 |
0.13 |
| chr19_36704891_36705475 | 0.06 |
ZNF146 |
zinc finger protein 146 |
321 |
0.51 |
| chr12_95043269_95044318 | 0.06 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
545 |
0.85 |
| chr8_22977984_22978135 | 0.06 |
TNFRSF10C |
tumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain |
17625 |
0.13 |
| chr17_58041207_58042016 | 0.06 |
RNFT1 |
ring finger protein, transmembrane 1 |
469 |
0.76 |
| chr11_118103487_118103638 | 0.06 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
7753 |
0.14 |
| chr14_107081796_107081947 | 0.06 |
IGHV4-59 |
immunoglobulin heavy variable 4-59 |
1854 |
0.07 |
| chr6_84936796_84937329 | 0.06 |
KIAA1009 |
KIAA1009 |
252 |
0.96 |
| chr1_76541245_76541396 | 0.06 |
ST6GALNAC3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
916 |
0.72 |
| chr19_1038413_1038573 | 0.06 |
AC011558.5 |
|
570 |
0.49 |
| chr8_8945449_8945788 | 0.06 |
ENSG00000239078 |
. |
15592 |
0.15 |
| chr17_57746374_57746525 | 0.06 |
CLTC |
clathrin, heavy chain (Hc) |
14818 |
0.17 |
| chr7_106504119_106504270 | 0.06 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1529 |
0.51 |
| chr15_101750389_101750540 | 0.06 |
CHSY1 |
chondroitin sulfate synthase 1 |
41673 |
0.14 |
| chr5_97903857_97904008 | 0.06 |
ENSG00000223053 |
. |
71003 |
0.13 |
| chr20_1945938_1946436 | 0.06 |
RP4-684O24.5 |
|
18295 |
0.2 |
| chr9_6552388_6552539 | 0.06 |
ENSG00000264530 |
. |
10369 |
0.19 |
| chr4_140376104_140376373 | 0.06 |
RP11-83A24.2 |
|
1050 |
0.38 |
| chr22_24644108_24644259 | 0.06 |
GGT5 |
gamma-glutamyltransferase 5 |
3073 |
0.21 |
| chr8_30601100_30602146 | 0.06 |
GSR |
glutathione reductase |
16180 |
0.2 |
| chr11_18404739_18405274 | 0.06 |
ENSG00000264603 |
. |
4328 |
0.14 |
| chr12_19391845_19391996 | 0.06 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
2062 |
0.39 |
| chr20_54997128_54997279 | 0.06 |
CASS4 |
Cas scaffolding protein family member 4 |
9886 |
0.13 |
| chr2_193100641_193100792 | 0.06 |
TMEFF2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
40281 |
0.22 |
| chr22_48594670_48594821 | 0.06 |
ENSG00000266508 |
. |
75431 |
0.13 |
| chr11_66079471_66080130 | 0.06 |
RP11-867G23.13 |
|
524 |
0.5 |
| chr4_164532971_164533122 | 0.06 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
1640 |
0.5 |
| chr4_174337851_174338086 | 0.06 |
SCRG1 |
stimulator of chondrogenesis 1 |
17281 |
0.15 |
| chr20_8113545_8114201 | 0.06 |
PLCB1 |
phospholipase C, beta 1 (phosphoinositide-specific) |
571 |
0.84 |
| chr19_35923693_35923844 | 0.06 |
FFAR2 |
free fatty acid receptor 2 |
15435 |
0.1 |
| chr19_47735595_47736220 | 0.06 |
BBC3 |
BCL2 binding component 3 |
116 |
0.95 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.1 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |