Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
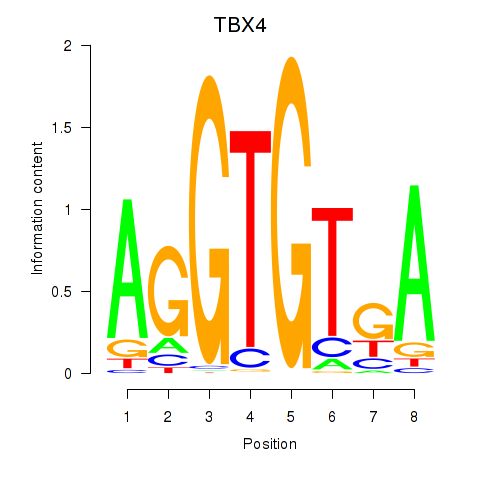
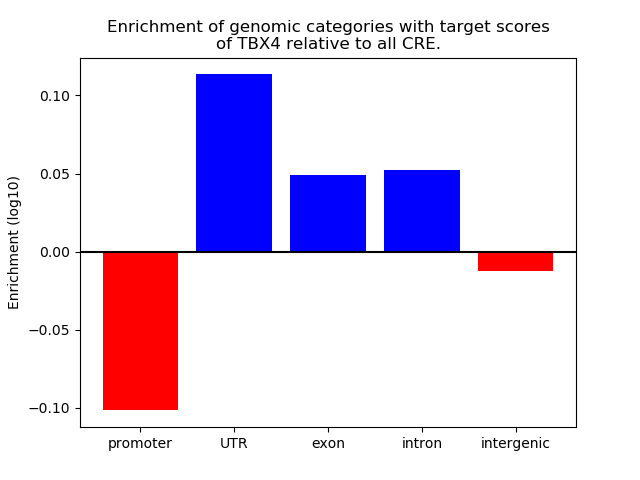
Results for TBX4
Z-value: 0.11
Transcription factors associated with TBX4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX4
|
ENSG00000121075.5 | T-box transcription factor 4 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_59530124_59530284 | TBX4 | 425 | 0.798140 | 0.39 | 2.9e-01 | Click! |
| chr17_59532021_59532172 | TBX4 | 1711 | 0.303618 | -0.33 | 3.8e-01 | Click! |
| chr17_59531737_59531888 | TBX4 | 1995 | 0.268096 | 0.27 | 4.9e-01 | Click! |
| chr17_59529188_59529339 | TBX4 | 502 | 0.747117 | -0.19 | 6.3e-01 | Click! |
| chr17_59529820_59529988 | TBX4 | 125 | 0.953028 | -0.09 | 8.2e-01 | Click! |
Activity of the TBX4 motif across conditions
Conditions sorted by the z-value of the TBX4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr7_158743544_158743695 | 0.02 |
ENSG00000231419 |
. |
57493 |
0.14 |
| chr4_75718841_75720131 | 0.02 |
BTC |
betacellulin |
410 |
0.91 |
| chr6_105388737_105389249 | 0.02 |
LIN28B |
lin-28 homolog B (C. elegans) |
15930 |
0.23 |
| chr2_243030793_243031816 | 0.02 |
AC093642.5 |
|
460 |
0.62 |
| chr22_36235496_36235773 | 0.02 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
631 |
0.82 |
| chr1_223302857_223303051 | 0.02 |
TLR5 |
toll-like receptor 5 |
5144 |
0.33 |
| chr15_81074624_81074775 | 0.02 |
KIAA1199 |
KIAA1199 |
2987 |
0.3 |
| chr8_93074610_93074813 | 0.02 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
480 |
0.89 |
| chr7_100424107_100424380 | 0.02 |
EPHB4 |
EPH receptor B4 |
494 |
0.66 |
| chr19_58769898_58770049 | 0.02 |
CTD-3138B18.6 |
|
8028 |
0.1 |
| chr1_110693132_110693795 | 0.02 |
SLC6A17 |
solute carrier family 6 (neutral amino acid transporter), member 17 |
355 |
0.81 |
| chr19_48102050_48102279 | 0.02 |
GLTSCR1 |
glioma tumor suppressor candidate region gene 1 |
9289 |
0.14 |
| chr4_169553568_169553728 | 0.02 |
PALLD |
palladin, cytoskeletal associated protein |
880 |
0.63 |
| chr10_15336245_15336418 | 0.02 |
RP11-25G10.2 |
|
56840 |
0.14 |
| chr6_11279481_11279645 | 0.02 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
46648 |
0.16 |
| chr5_36744281_36744616 | 0.02 |
CTD-2353F22.1 |
|
19151 |
0.27 |
| chr22_46482810_46483184 | 0.02 |
FLJ27365 |
hsa-mir-4763 |
1114 |
0.31 |
| chr1_205648469_205649627 | 0.02 |
SLC45A3 |
solute carrier family 45, member 3 |
539 |
0.74 |
| chr16_3740619_3740949 | 0.02 |
TRAP1 |
TNF receptor-associated protein 1 |
350 |
0.86 |
| chr3_87033354_87033505 | 0.02 |
VGLL3 |
vestigial like 3 (Drosophila) |
6423 |
0.34 |
| chr9_127143624_127143775 | 0.02 |
ENSG00000264237 |
. |
30889 |
0.15 |
| chr2_74606600_74607395 | 0.01 |
DCTN1 |
dynactin 1 |
413 |
0.73 |
| chr11_9511154_9511374 | 0.01 |
ZNF143 |
zinc finger protein 143 |
22780 |
0.14 |
| chr11_118851261_118851557 | 0.01 |
FOXR1 |
forkhead box R1 |
1920 |
0.13 |
| chr8_131032307_131032560 | 0.01 |
FAM49B |
family with sequence similarity 49, member B |
3058 |
0.23 |
| chr15_75943747_75944181 | 0.01 |
SNX33 |
sorting nexin 33 |
1867 |
0.18 |
| chr19_676845_677141 | 0.01 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
601 |
0.56 |
| chr12_111843881_111845308 | 0.01 |
SH2B3 |
SH2B adaptor protein 3 |
842 |
0.62 |
| chr11_61448104_61448359 | 0.01 |
DAGLA |
diacylglycerol lipase, alpha |
326 |
0.89 |
| chr7_100773540_100774051 | 0.01 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
3416 |
0.13 |
| chr13_40175117_40175459 | 0.01 |
LHFP |
lipoma HMGIC fusion partner |
2020 |
0.37 |
| chr8_119068765_119068981 | 0.01 |
EXT1 |
exostosin glycosyltransferase 1 |
53780 |
0.18 |
| chr6_28058943_28059326 | 0.01 |
ZSCAN12P1 |
zinc finger and SCAN domain containing 12 pseudogene 1 |
1071 |
0.38 |
| chr19_47735595_47736220 | 0.01 |
BBC3 |
BCL2 binding component 3 |
116 |
0.95 |
| chr21_17489674_17489828 | 0.01 |
ENSG00000252273 |
. |
81922 |
0.11 |
| chr10_104465015_104465218 | 0.01 |
ARL3 |
ADP-ribosylation factor-like 3 |
9048 |
0.15 |
| chr15_48013744_48013895 | 0.01 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
3133 |
0.37 |
| chr2_16832297_16832583 | 0.01 |
FAM49A |
family with sequence similarity 49, member A |
14656 |
0.3 |
| chr7_20391312_20391720 | 0.01 |
CTA-293F17.1 |
|
20146 |
0.18 |
| chr9_109624062_109624226 | 0.01 |
ZNF462 |
zinc finger protein 462 |
1234 |
0.53 |
| chr14_105452072_105453062 | 0.01 |
C14orf79 |
chromosome 14 open reading frame 79 |
437 |
0.8 |
| chr17_38500769_38502147 | 0.01 |
RARA |
retinoic acid receptor, alpha |
35 |
0.95 |
| chr10_111834916_111835067 | 0.01 |
ADD3 |
adducin 3 (gamma) |
67269 |
0.1 |
| chr4_74847354_74847841 | 0.01 |
PF4 |
platelet factor 4 |
244 |
0.89 |
| chr7_147097205_147097356 | 0.01 |
ENSG00000221442 |
. |
22067 |
0.28 |
| chr16_72961288_72961637 | 0.01 |
ENSG00000221799 |
. |
57294 |
0.12 |
| chr1_206687074_206687508 | 0.01 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
6412 |
0.15 |
| chr3_190415622_190416004 | 0.01 |
ENSG00000223117 |
. |
55861 |
0.15 |
| chr4_41001068_41001219 | 0.01 |
ENSG00000201736 |
. |
8972 |
0.21 |
| chr2_239869839_239870212 | 0.01 |
ENSG00000211566 |
. |
24681 |
0.17 |
| chr2_216288358_216288696 | 0.01 |
FN1 |
fibronectin 1 |
12263 |
0.21 |
| chr18_53006641_53006980 | 0.01 |
TCF4 |
transcription factor 4 |
12418 |
0.28 |
| chr10_126745527_126745695 | 0.01 |
ENSG00000264572 |
. |
24172 |
0.19 |
| chr11_72975754_72976333 | 0.01 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
232 |
0.91 |
| chr3_151046365_151046540 | 0.01 |
P2RY13 |
purinergic receptor P2Y, G-protein coupled, 13 |
884 |
0.57 |
| chr19_12016806_12016962 | 0.01 |
ZNF69 |
zinc finger protein 69 |
18214 |
0.1 |
| chrX_53145254_53145405 | 0.01 |
ENSG00000251707 |
. |
14334 |
0.16 |
| chr6_137242277_137243470 | 0.01 |
SLC35D3 |
solute carrier family 35, member D3 |
529 |
0.78 |
| chr1_114522595_114522893 | 0.01 |
OLFML3 |
olfactomedin-like 3 |
49 |
0.97 |
| chr1_235097453_235097604 | 0.01 |
ENSG00000239690 |
. |
57595 |
0.14 |
| chr5_88171366_88171720 | 0.01 |
MEF2C |
myocyte enhancer factor 2C |
2251 |
0.36 |
| chr9_99429026_99429186 | 0.01 |
AAED1 |
AhpC/TSA antioxidant enzyme domain containing 1 |
11521 |
0.24 |
| chr12_6873296_6873504 | 0.01 |
PTMS |
parathymosin |
2141 |
0.13 |
| chr1_15527398_15527585 | 0.01 |
C1orf195 |
chromosome 1 open reading frame 195 |
29678 |
0.16 |
| chr19_37129764_37129915 | 0.01 |
ZNF461 |
zinc finger protein 461 |
27886 |
0.11 |
| chr5_150022452_150022802 | 0.01 |
SYNPO |
synaptopodin |
2387 |
0.23 |
| chr21_35892920_35893071 | 0.01 |
RCAN1 |
regulator of calcineurin 1 |
3592 |
0.23 |
| chr1_109045091_109045361 | 0.01 |
NBPF6 |
neuroblastoma breakpoint family, member 6 |
52283 |
0.14 |
| chr13_106828289_106828440 | 0.01 |
ENSG00000222682 |
. |
20525 |
0.25 |
| chr11_74561071_74561376 | 0.01 |
ENSG00000242999 |
. |
4268 |
0.2 |
| chr14_31343280_31344466 | 0.01 |
COCH |
cochlin |
78 |
0.97 |
| chr18_26205812_26205963 | 0.01 |
ENSG00000251719 |
. |
422697 |
0.01 |
| chr3_157248805_157248956 | 0.01 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
2515 |
0.34 |
| chr15_40697977_40698740 | 0.01 |
IVD |
isovaleryl-CoA dehydrogenase |
83 |
0.94 |
| chr13_113597266_113598290 | 0.01 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
24979 |
0.15 |
| chr8_126322078_126322527 | 0.01 |
ENSG00000242170 |
. |
39456 |
0.16 |
| chr18_55448645_55448916 | 0.01 |
ATP8B1 |
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
21547 |
0.18 |
| chr8_8182381_8182560 | 0.01 |
SGK223 |
Tyrosine-protein kinase SgK223 |
56787 |
0.12 |
| chr4_186456818_186456988 | 0.01 |
PDLIM3 |
PDZ and LIM domain 3 |
137 |
0.96 |
| chr22_43005089_43005553 | 0.01 |
POLDIP3 |
polymerase (DNA-directed), delta interacting protein 3 |
5542 |
0.12 |
| chr11_27743435_27743805 | 0.01 |
BDNF |
brain-derived neurotrophic factor |
15 |
0.99 |
| chr10_10834416_10834694 | 0.01 |
CELF2 |
CUGBP, Elav-like family member 2 |
212704 |
0.02 |
| chr8_96127862_96128066 | 0.01 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
18068 |
0.16 |
| chr10_129704991_129705210 | 0.01 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
225 |
0.94 |
| chr3_162003818_162003969 | 0.01 |
OTOL1 |
otolin 1 |
789297 |
0.0 |
| chr21_37305025_37305176 | 0.01 |
FKSG68 |
|
34373 |
0.16 |
| chr16_5006063_5006260 | 0.01 |
SEC14L5 |
SEC14-like 5 (S. cerevisiae) |
2157 |
0.23 |
| chr15_62737317_62737509 | 0.01 |
TLN2 |
talin 2 |
116151 |
0.06 |
| chr1_21669455_21669746 | 0.01 |
ECE1 |
endothelin converting enzyme 1 |
2397 |
0.33 |
| chr22_43685292_43685751 | 0.01 |
Z99756.1 |
|
13560 |
0.18 |
| chr19_14605464_14605615 | 0.01 |
GIPC1 |
GIPC PDZ domain containing family, member 1 |
1405 |
0.26 |
| chr7_28387506_28387665 | 0.01 |
CREB5 |
cAMP responsive element binding protein 5 |
48645 |
0.18 |
| chr10_13745153_13745512 | 0.01 |
ENSG00000222235 |
. |
1646 |
0.27 |
| chr2_112456971_112457256 | 0.01 |
ENSG00000266063 |
. |
71598 |
0.12 |
| chr9_21677527_21678485 | 0.01 |
ENSG00000244230 |
. |
21307 |
0.21 |
| chr3_157157796_157157947 | 0.01 |
PTX3 |
pentraxin 3, long |
3293 |
0.33 |
| chrX_67867131_67867291 | 0.01 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
300 |
0.94 |
| chr9_127999236_127999759 | 0.01 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
4112 |
0.17 |
| chr3_8561489_8561767 | 0.01 |
LMCD1 |
LIM and cysteine-rich domains 1 |
17241 |
0.2 |
| chr15_86095041_86095192 | 0.01 |
AKAP13 |
A kinase (PRKA) anchor protein 13 |
3561 |
0.24 |
| chr19_3397384_3397535 | 0.01 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
30875 |
0.13 |
| chr19_16179581_16179939 | 0.01 |
TPM4 |
tropomyosin 4 |
1250 |
0.44 |
| chr15_52860024_52861356 | 0.01 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
339 |
0.87 |
| chr20_42667406_42667585 | 0.01 |
TOX2 |
TOX high mobility group box family member 2 |
15496 |
0.21 |
| chr2_237570334_237570575 | 0.01 |
ACKR3 |
atypical chemokine receptor 3 |
92170 |
0.09 |
| chr16_29836314_29836619 | 0.01 |
MVP |
major vault protein |
3820 |
0.08 |
| chr11_6340007_6340271 | 0.01 |
PRKCDBP |
protein kinase C, delta binding protein |
1623 |
0.33 |
| chr2_177361445_177361596 | 0.01 |
ENSG00000221304 |
. |
113216 |
0.06 |
| chr6_36592551_36592702 | 0.01 |
ENSG00000263894 |
. |
2337 |
0.22 |
| chr7_15503276_15503427 | 0.01 |
AGMO |
alkylglycerol monooxygenase |
97504 |
0.09 |
| chr11_74560843_74561007 | 0.01 |
ENSG00000242999 |
. |
3970 |
0.2 |
| chr1_6803789_6803981 | 0.01 |
ENSG00000239166 |
. |
8894 |
0.17 |
| chr7_120954228_120954379 | 0.01 |
WNT16 |
wingless-type MMTV integration site family, member 16 |
11118 |
0.27 |
| chr9_77725905_77726056 | 0.01 |
OSTF1 |
osteoclast stimulating factor 1 |
22521 |
0.16 |
| chr17_80970427_80970618 | 0.01 |
RP11-1197K16.2 |
|
31564 |
0.16 |
| chr17_2143243_2143467 | 0.01 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
130 |
0.93 |
| chr5_176798199_176798886 | 0.01 |
RGS14 |
regulator of G-protein signaling 14 |
4598 |
0.11 |
| chr10_60515616_60516529 | 0.01 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
37223 |
0.22 |
| chr19_12016356_12016507 | 0.01 |
ZNF69 |
zinc finger protein 69 |
17761 |
0.1 |
| chrX_19701431_19701593 | 0.01 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
12396 |
0.3 |
| chr2_43360552_43360703 | 0.01 |
ENSG00000207087 |
. |
41995 |
0.18 |
| chr1_174081654_174081874 | 0.01 |
RABGAP1L |
RAB GTPase activating protein 1-like |
46784 |
0.14 |
| chr3_43795580_43795774 | 0.01 |
ABHD5 |
abhydrolase domain containing 5 |
42478 |
0.18 |
| chr1_25870143_25871217 | 0.01 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
609 |
0.77 |
| chr8_30601100_30602146 | 0.01 |
GSR |
glutathione reductase |
16180 |
0.2 |
| chr16_75654262_75654413 | 0.01 |
ADAT1 |
adenosine deaminase, tRNA-specific 1 |
2344 |
0.22 |
| chr3_43298080_43298231 | 0.01 |
SNRK |
SNF related kinase |
29849 |
0.16 |
| chr3_149266255_149266548 | 0.01 |
ENSG00000252581 |
. |
22983 |
0.17 |
| chr3_197036778_197036943 | 0.01 |
DLG1 |
discs, large homolog 1 (Drosophila) |
10689 |
0.16 |
| chr4_113220908_113221059 | 0.01 |
ALPK1 |
alpha-kinase 1 |
2447 |
0.25 |
| chr4_160096780_160097268 | 0.01 |
ENSG00000264105 |
. |
46978 |
0.13 |
| chr2_106010267_106010729 | 0.01 |
FHL2 |
four and a half LIM domains 2 |
2744 |
0.27 |
| chr12_118491012_118491163 | 0.01 |
WSB2 |
WD repeat and SOCS box containing 2 |
678 |
0.71 |
| chr19_53668202_53668353 | 0.01 |
CTD-2245F17.2 |
|
2596 |
0.18 |
| chr10_128352295_128352662 | 0.01 |
C10orf90 |
chromosome 10 open reading frame 90 |
6537 |
0.3 |
| chr1_15297301_15297452 | 0.01 |
KAZN |
kazrin, periplakin interacting protein |
24961 |
0.26 |
| chr5_76475983_76476746 | 0.01 |
PDE8B |
phosphodiesterase 8B |
29910 |
0.15 |
| chr7_106263871_106264022 | 0.01 |
CTB-111H14.1 |
|
16408 |
0.24 |
| chr12_124773798_124773955 | 0.01 |
FAM101A |
family with sequence similarity 101, member A |
166 |
0.97 |
| chr4_169984004_169984155 | 0.01 |
RP11-483A20.3 |
|
52498 |
0.12 |
| chr3_189206116_189206267 | 0.01 |
TP63 |
tumor protein p63 |
143025 |
0.05 |
| chr10_28537339_28537590 | 0.01 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
9798 |
0.27 |
| chr11_70246165_70246316 | 0.01 |
CTTN |
cortactin |
1593 |
0.25 |
| chr6_121760674_121760877 | 0.01 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
3937 |
0.22 |
| chr18_60920514_60920680 | 0.01 |
ENSG00000238988 |
. |
58699 |
0.1 |
| chr9_6970541_6970692 | 0.01 |
KDM4C |
lysine (K)-specific demethylase 4C |
43172 |
0.19 |
| chr17_70636816_70636967 | 0.01 |
ENSG00000200783 |
. |
23400 |
0.24 |
| chr16_87250830_87251124 | 0.01 |
RP11-899L11.3 |
|
1456 |
0.45 |
| chr20_30149403_30149560 | 0.01 |
HM13-IT1 |
HM13 intronic transcript 1 (non-protein coding) |
1488 |
0.28 |
| chr20_49154846_49154997 | 0.01 |
ENSG00000239742 |
. |
19614 |
0.13 |
| chr15_68575595_68575746 | 0.01 |
FEM1B |
fem-1 homolog b (C. elegans) |
3373 |
0.24 |
| chr15_60719892_60720043 | 0.01 |
ANXA2 |
annexin A2 |
24885 |
0.19 |
| chrX_19687578_19687729 | 0.01 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
827 |
0.76 |
| chr5_14268280_14268580 | 0.01 |
TRIO |
trio Rho guanine nucleotide exchange factor |
22656 |
0.28 |
| chr18_74779349_74779519 | 0.01 |
MBP |
myelin basic protein |
37783 |
0.2 |
| chr1_119521934_119522435 | 0.01 |
TBX15 |
T-box 15 |
8244 |
0.28 |
| chr1_12579130_12579281 | 0.01 |
ENSG00000239149 |
. |
11905 |
0.19 |
| chr18_70514172_70514326 | 0.01 |
ENSG00000263750 |
. |
6368 |
0.24 |
| chr4_15907365_15907907 | 0.01 |
FGFBP1 |
fibroblast growth factor binding protein 1 |
32335 |
0.17 |
| chr11_121971856_121972438 | 0.01 |
RP11-166D19.1 |
|
247 |
0.88 |
| chr3_99964720_99964871 | 0.01 |
TBC1D23 |
TBC1 domain family, member 23 |
15049 |
0.17 |
| chr9_115629928_115630101 | 0.01 |
RP11-408O19.5 |
|
17424 |
0.18 |
| chr6_112545177_112545328 | 0.01 |
RP11-506B6.6 |
|
12042 |
0.18 |
| chr8_23201528_23201679 | 0.01 |
ENSG00000253837 |
. |
7882 |
0.17 |
| chr16_66555077_66555228 | 0.01 |
TK2 |
thymidine kinase 2, mitochondrial |
7466 |
0.11 |
| chr14_69481156_69481307 | 0.01 |
ACTN1-AS1 |
ACTN1 antisense RNA 1 |
34473 |
0.17 |
| chr12_40635540_40635691 | 0.01 |
LRRK2 |
leucine-rich repeat kinase 2 |
16739 |
0.22 |
| chr3_149297214_149297365 | 0.01 |
WWTR1 |
WW domain containing transcription regulator 1 |
3250 |
0.27 |
| chr6_144461046_144461197 | 0.01 |
STX11 |
syntaxin 11 |
10542 |
0.24 |
| chr8_68304896_68305241 | 0.01 |
ARFGEF1 |
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
49156 |
0.18 |
| chr13_80615426_80615577 | 0.01 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
298293 |
0.01 |
| chr3_177055525_177055734 | 0.01 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
140368 |
0.05 |
| chr2_176432418_176432569 | 0.01 |
ENSG00000221347 |
. |
237392 |
0.02 |
| chr19_50009986_50010137 | 0.01 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
5475 |
0.06 |
| chrX_41243634_41243785 | 0.01 |
ENSG00000264573 |
. |
38580 |
0.14 |
| chr3_55518754_55518905 | 0.01 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
2502 |
0.32 |
| chr16_57229031_57229182 | 0.01 |
RSPRY1 |
ring finger and SPRY domain containing 1 |
8861 |
0.14 |
| chr15_89636866_89637017 | 0.01 |
ABHD2 |
abhydrolase domain containing 2 |
5280 |
0.19 |
| chr10_81933461_81933612 | 0.01 |
ANXA11 |
annexin A11 |
759 |
0.68 |
| chr1_98685801_98685961 | 0.01 |
ENSG00000221777 |
. |
153060 |
0.04 |
| chr3_43020505_43020694 | 0.01 |
FAM198A |
family with sequence similarity 198, member A |
174 |
0.95 |
| chr6_75992353_75992507 | 0.01 |
TMEM30A |
transmembrane protein 30A |
1597 |
0.4 |
| chr7_19155354_19155672 | 0.01 |
TWIST1 |
twist family bHLH transcription factor 1 |
1782 |
0.28 |
| chr10_14799506_14799657 | 0.01 |
FAM107B |
family with sequence similarity 107, member B |
17315 |
0.21 |
| chr21_27762391_27762594 | 0.01 |
AP001596.6 |
|
37 |
0.98 |
| chr9_88401280_88401431 | 0.01 |
AGTPBP1 |
ATP/GTP binding protein 1 |
44411 |
0.21 |
| chr16_69139287_69139438 | 0.01 |
HAS3 |
hyaluronan synthase 3 |
105 |
0.96 |
| chr1_224076105_224076307 | 0.01 |
TP53BP2 |
tumor protein p53 binding protein, 2 |
42532 |
0.17 |
| chr1_110407367_110407518 | 0.01 |
RP11-195M16.1 |
|
21444 |
0.16 |
| chr5_167004315_167004466 | 0.01 |
CTB-78F1.1 |
|
83096 |
0.1 |
| chr9_108007124_108008196 | 0.01 |
SLC44A1 |
solute carrier family 44 (choline transporter), member 1 |
757 |
0.78 |
| chr8_13265944_13266113 | 0.01 |
RP11-10C8.2 |
|
69945 |
0.11 |
| chr22_31039873_31040024 | 0.01 |
SLC35E4 |
solute carrier family 35, member E4 |
7438 |
0.13 |
| chr19_16555208_16555359 | 0.01 |
CTD-2013N17.4 |
|
3526 |
0.16 |
| chr5_60745627_60745803 | 0.01 |
ENSG00000223200 |
. |
107395 |
0.07 |
| chr19_46046997_46047180 | 0.01 |
VASP |
vasodilator-stimulated phosphoprotein |
36339 |
0.08 |
| chr4_8421351_8421502 | 0.01 |
ACOX3 |
acyl-CoA oxidase 3, pristanoyl |
8762 |
0.17 |
| chr1_38022972_38023123 | 0.01 |
DNALI1 |
dynein, axonemal, light intermediate chain 1 |
440 |
0.76 |
| chr7_3957020_3957171 | 0.01 |
SDK1 |
sidekick cell adhesion molecule 1 |
33457 |
0.25 |
| chr19_44179251_44179402 | 0.01 |
PLAUR |
plasminogen activator, urokinase receptor |
4627 |
0.11 |
Network of associatons between targets according to the STRING database.