Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
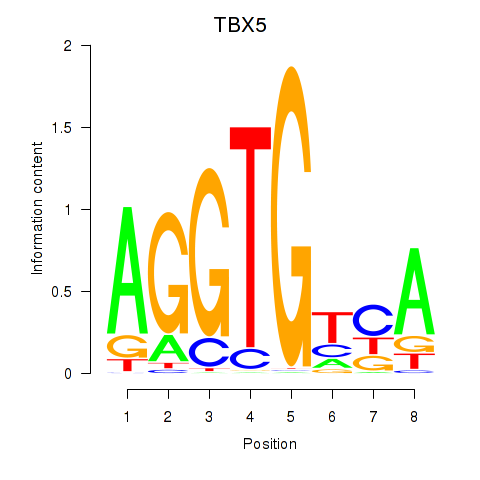
Results for TBX5
Z-value: 0.92
Transcription factors associated with TBX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TBX5
|
ENSG00000089225.15 | T-box transcription factor 5 |
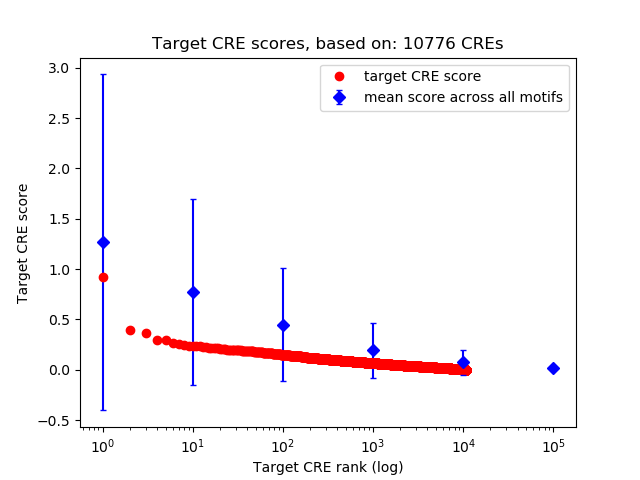
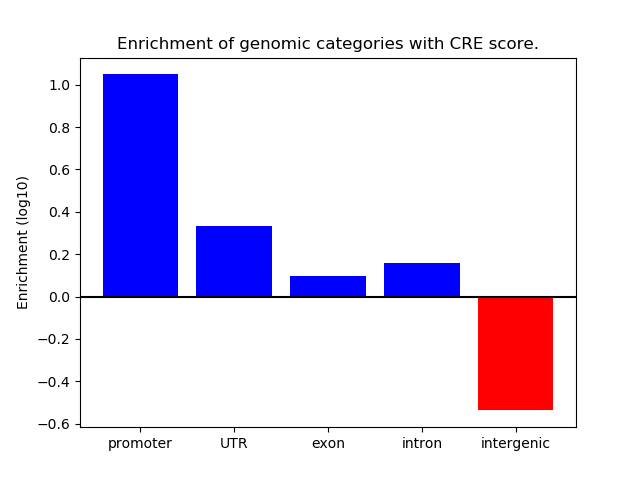
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_114744833_114744984 | TBX5 | 96795 | 0.080510 | -0.77 | 1.5e-02 | Click! |
| chr12_114757258_114757409 | TBX5 | 84370 | 0.095488 | -0.50 | 1.7e-01 | Click! |
| chr12_114660353_114660504 | TBX5 | 181275 | 0.030477 | -0.39 | 3.0e-01 | Click! |
| chr12_114839312_114839463 | TBX5 | 2316 | 0.342807 | -0.30 | 4.3e-01 | Click! |
| chr12_114700866_114701017 | TBX5 | 140762 | 0.046593 | -0.28 | 4.7e-01 | Click! |
Activity of the TBX5 motif across conditions
Conditions sorted by the z-value of the TBX5 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
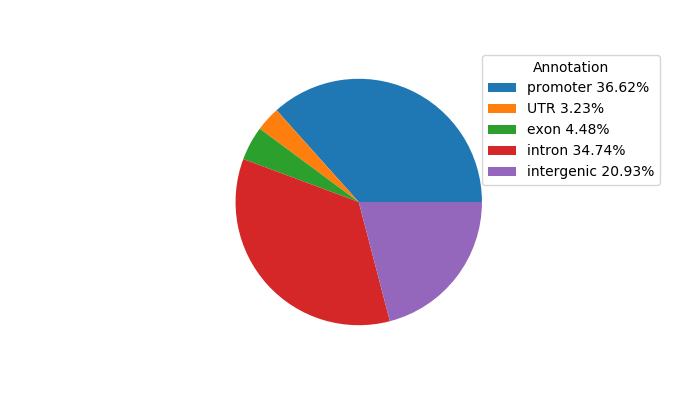
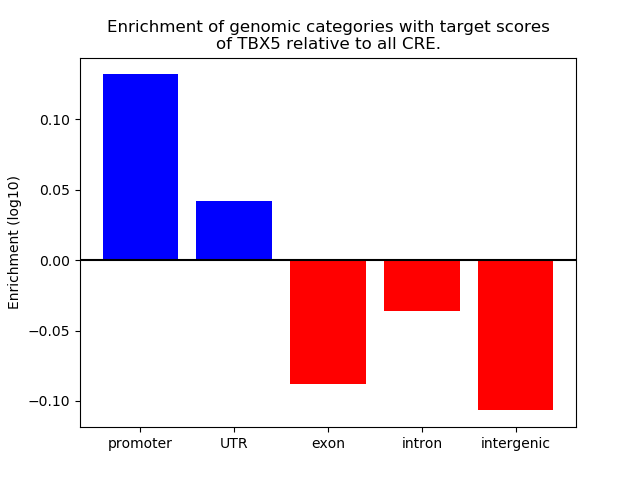
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_640288_641115 | 0.92 |
RAB40C |
RAB40C, member RAS oncogene family |
369 |
0.68 |
| chr16_641141_641376 | 0.40 |
RAB40C |
RAB40C, member RAS oncogene family |
926 |
0.31 |
| chr5_176728326_176728485 | 0.36 |
RAB24 |
RAB24, member RAS oncogene family |
2302 |
0.17 |
| chr1_230262959_230263110 | 0.30 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
60016 |
0.13 |
| chr4_100738015_100738341 | 0.29 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
175 |
0.97 |
| chr2_219031294_219031758 | 0.27 |
CXCR1 |
chemokine (C-X-C motif) receptor 1 |
192 |
0.92 |
| chr1_181002597_181002748 | 0.25 |
MR1 |
major histocompatibility complex, class I-related |
395 |
0.84 |
| chr11_47398844_47399090 | 0.24 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
975 |
0.36 |
| chr2_10472002_10472153 | 0.24 |
HPCAL1 |
hippocalcin-like 1 |
28251 |
0.15 |
| chr4_153599291_153599457 | 0.24 |
TMEM154 |
transmembrane protein 154 |
1790 |
0.38 |
| chrX_128913998_128914362 | 0.23 |
SASH3 |
SAM and SH3 domain containing 3 |
220 |
0.94 |
| chr19_2289358_2289509 | 0.23 |
LINGO3 |
leucine rich repeat and Ig domain containing 3 |
2590 |
0.13 |
| chr22_37317841_37317992 | 0.23 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
260 |
0.88 |
| chr8_48515171_48515322 | 0.23 |
SPIDR |
scaffolding protein involved in DNA repair |
56980 |
0.12 |
| chr12_54693479_54693635 | 0.22 |
NFE2 |
nuclear factor, erythroid 2 |
1242 |
0.21 |
| chr7_37683935_37684086 | 0.22 |
GPR141 |
G protein-coupled receptor 141 |
39390 |
0.17 |
| chr22_24038685_24038889 | 0.22 |
RGL4 |
ral guanine nucleotide dissociation stimulator-like 4 |
116 |
0.94 |
| chr12_122250872_122251023 | 0.22 |
SETD1B |
SET domain containing 1B |
8309 |
0.15 |
| chr22_50050359_50050996 | 0.21 |
C22orf34 |
chromosome 22 open reading frame 34 |
401 |
0.87 |
| chr19_2089521_2089711 | 0.21 |
MOB3A |
MOB kinase activator 3A |
559 |
0.63 |
| chr18_72256195_72256346 | 0.21 |
ZNF407 |
zinc finger protein 407 |
8836 |
0.21 |
| chr13_114261602_114261753 | 0.20 |
TFDP1 |
transcription factor Dp-1 |
21936 |
0.18 |
| chr6_160374023_160374174 | 0.20 |
IGF2R |
insulin-like growth factor 2 receptor |
16033 |
0.2 |
| chr2_162101279_162101855 | 0.20 |
AC009299.2 |
|
6106 |
0.21 |
| chr8_23416478_23416629 | 0.20 |
AC051642.1 |
|
4098 |
0.19 |
| chr1_26870560_26870711 | 0.20 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
1032 |
0.45 |
| chr3_126248153_126248304 | 0.20 |
CHST13 |
carbohydrate (chondroitin 4) sulfotransferase 13 |
5102 |
0.16 |
| chr15_41195656_41195807 | 0.20 |
VPS18 |
vacuolar protein sorting 18 homolog (S. cerevisiae) |
9066 |
0.11 |
| chr14_69413231_69413479 | 0.19 |
ACTN1 |
actinin, alpha 1 |
910 |
0.65 |
| chr3_71773124_71773375 | 0.19 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
1277 |
0.5 |
| chr14_61790070_61790226 | 0.19 |
ENSG00000247287 |
. |
612 |
0.51 |
| chr10_125852855_125853389 | 0.19 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
84 |
0.98 |
| chr7_45017874_45018185 | 0.19 |
MYO1G |
myosin IG |
668 |
0.61 |
| chr17_29816015_29816298 | 0.19 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
1030 |
0.45 |
| chr9_134903947_134904098 | 0.19 |
MED27 |
mediator complex subunit 27 |
51231 |
0.16 |
| chr20_46294380_46294531 | 0.19 |
SULF2 |
sulfatase 2 |
452 |
0.85 |
| chr3_184053343_184053608 | 0.19 |
FAM131A |
family with sequence similarity 131, member A |
239 |
0.85 |
| chr10_82378763_82378914 | 0.19 |
SH2D4B |
SH2 domain containing 4B |
78263 |
0.1 |
| chr12_53772533_53773270 | 0.19 |
SP1 |
Sp1 transcription factor |
1059 |
0.4 |
| chr17_3814974_3815160 | 0.19 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
4727 |
0.18 |
| chr11_46378846_46378997 | 0.19 |
DGKZ |
diacylglycerol kinase, zeta |
4224 |
0.16 |
| chr3_14472654_14472930 | 0.18 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
1386 |
0.5 |
| chr8_56795950_56796101 | 0.18 |
LYN |
v-yes-1 Yamaguchi sarcoma viral related oncogene homolog |
3631 |
0.18 |
| chr1_244488017_244488168 | 0.18 |
C1orf100 |
chromosome 1 open reading frame 100 |
27845 |
0.22 |
| chr17_43302327_43302665 | 0.18 |
CTD-2020K17.1 |
|
2907 |
0.13 |
| chr1_174934321_174935265 | 0.18 |
RABGAP1L |
RAB GTPase activating protein 1-like |
888 |
0.52 |
| chr8_134086172_134086360 | 0.18 |
SLA |
Src-like-adaptor |
13663 |
0.23 |
| chr9_129090297_129090448 | 0.18 |
MVB12B |
multivesicular body subunit 12B |
1244 |
0.58 |
| chr13_111317307_111317458 | 0.18 |
CARS2 |
cysteinyl-tRNA synthetase 2, mitochondrial (putative) |
11989 |
0.21 |
| chr2_263631_263782 | 0.18 |
SH3YL1 |
SH3 and SYLF domain containing 1 |
326 |
0.81 |
| chr10_111897137_111897288 | 0.18 |
MXI1 |
MAX interactor 1, dimerization protein |
70151 |
0.1 |
| chr1_28452985_28453161 | 0.18 |
ENSG00000253005 |
. |
16279 |
0.12 |
| chr6_38019767_38019918 | 0.18 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
122107 |
0.06 |
| chr1_154977279_154977430 | 0.18 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
2059 |
0.13 |
| chr5_80655477_80655628 | 0.18 |
ACOT12 |
acyl-CoA thioesterase 12 |
34410 |
0.17 |
| chr8_145870561_145870712 | 0.17 |
ARHGAP39 |
Rho GTPase activating protein 39 |
39435 |
0.09 |
| chr20_5100852_5101135 | 0.17 |
PCNA |
proliferating cell nuclear antigen |
321 |
0.77 |
| chr14_71807680_71807831 | 0.17 |
RP1-261D10.2 |
|
19224 |
0.19 |
| chr17_17524925_17525076 | 0.17 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
29978 |
0.13 |
| chr1_45284842_45285269 | 0.17 |
ENSG00000202444 |
. |
614 |
0.49 |
| chr12_50350018_50350169 | 0.17 |
RP11-469H8.8 |
|
185 |
0.9 |
| chr19_18682141_18682437 | 0.17 |
UBA52 |
ubiquitin A-52 residue ribosomal protein fusion product 1 |
251 |
0.8 |
| chr7_8010063_8010681 | 0.17 |
AC006042.7 |
|
838 |
0.45 |
| chr4_40202563_40202865 | 0.17 |
RHOH |
ras homolog family member H |
750 |
0.7 |
| chr1_113589029_113589180 | 0.17 |
LRIG2 |
leucine-rich repeats and immunoglobulin-like domains 2 |
26727 |
0.22 |
| chr9_101881133_101881552 | 0.17 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
8715 |
0.21 |
| chr5_180231596_180231747 | 0.17 |
MGAT1 |
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
748 |
0.63 |
| chr12_123372500_123372784 | 0.17 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
2064 |
0.29 |
| chr1_205290973_205291124 | 0.17 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
165 |
0.95 |
| chr2_113593759_113594336 | 0.16 |
IL1B |
interleukin 1, beta |
36 |
0.97 |
| chr1_161595583_161595734 | 0.16 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
5164 |
0.13 |
| chr10_134823331_134823482 | 0.16 |
GPR123 |
G protein-coupled receptor 123 |
61027 |
0.11 |
| chr6_35271134_35271290 | 0.16 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
5583 |
0.2 |
| chr10_121351719_121352411 | 0.16 |
TIAL1 |
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
3942 |
0.27 |
| chr22_19675804_19675955 | 0.16 |
SEPT5 |
septin 5 |
26108 |
0.16 |
| chr7_101929391_101929542 | 0.16 |
SH2B2 |
SH2B adaptor protein 2 |
1014 |
0.42 |
| chr9_74398132_74398283 | 0.16 |
TMEM2 |
transmembrane protein 2 |
14407 |
0.27 |
| chr12_51782252_51782403 | 0.16 |
GALNT6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) |
2756 |
0.2 |
| chr11_61739742_61739933 | 0.16 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
4384 |
0.15 |
| chr4_153595488_153595639 | 0.16 |
TMEM154 |
transmembrane protein 154 |
5601 |
0.22 |
| chr13_110441346_110441497 | 0.16 |
IRS2 |
insulin receptor substrate 2 |
2506 |
0.39 |
| chr20_56195172_56195527 | 0.16 |
ZBP1 |
Z-DNA binding protein 1 |
101 |
0.98 |
| chrX_67900625_67900776 | 0.16 |
STARD8 |
StAR-related lipid transfer (START) domain containing 8 |
12793 |
0.29 |
| chr17_73959756_73959907 | 0.16 |
ACOX1 |
acyl-CoA oxidase 1, palmitoyl |
14218 |
0.09 |
| chr2_238583229_238583380 | 0.16 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
17484 |
0.2 |
| chr8_82022969_82023806 | 0.16 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
916 |
0.72 |
| chr19_42130849_42131075 | 0.16 |
CEACAM4 |
carcinoembryonic antigen-related cell adhesion molecule 4 |
2480 |
0.23 |
| chr12_109084399_109085395 | 0.16 |
CORO1C |
coronin, actin binding protein, 1C |
5281 |
0.17 |
| chr21_44466329_44466480 | 0.16 |
CBS |
cystathionine-beta-synthase |
12647 |
0.18 |
| chr1_160616915_160617313 | 0.16 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
29 |
0.97 |
| chr3_19188647_19189167 | 0.16 |
KCNH8 |
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
1039 |
0.7 |
| chr10_133608786_133608937 | 0.16 |
AL450307.1 |
Uncharacterized protein; cDNA FLJ46300 fis, clone TESTI4035989 |
13674 |
0.28 |
| chr1_9689695_9689868 | 0.16 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
22009 |
0.15 |
| chr19_33690657_33690808 | 0.15 |
LRP3 |
low density lipoprotein receptor-related protein 3 |
5242 |
0.14 |
| chr19_16477276_16478284 | 0.15 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
5016 |
0.17 |
| chr5_135348018_135348169 | 0.15 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
16491 |
0.2 |
| chr6_27635076_27635227 | 0.15 |
ENSG00000238648 |
. |
35963 |
0.14 |
| chr1_153488579_153488730 | 0.15 |
ENSG00000263841 |
. |
15414 |
0.08 |
| chr20_61203145_61203296 | 0.15 |
ENSG00000207764 |
. |
41101 |
0.09 |
| chr20_30154465_30154616 | 0.15 |
HM13-IT1 |
HM13 intronic transcript 1 (non-protein coding) |
3571 |
0.15 |
| chr1_154981837_154981988 | 0.15 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
5012 |
0.08 |
| chr12_105076123_105076407 | 0.15 |
ENSG00000264295 |
. |
90854 |
0.08 |
| chr11_47382738_47382889 | 0.15 |
MYBPC3 |
myosin binding protein C, cardiac |
8560 |
0.1 |
| chr10_70090873_70091608 | 0.15 |
HNRNPH3 |
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
592 |
0.65 |
| chr2_68592484_68592752 | 0.15 |
AC015969.3 |
|
98 |
0.77 |
| chr9_95728150_95728303 | 0.15 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
1983 |
0.37 |
| chr19_3135427_3135578 | 0.15 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
689 |
0.53 |
| chr12_54763014_54763165 | 0.15 |
GPR84 |
G protein-coupled receptor 84 |
4818 |
0.1 |
| chr8_67973634_67974505 | 0.15 |
COPS5 |
COP9 signalosome subunit 5 |
483 |
0.5 |
| chr9_100954099_100954367 | 0.15 |
CORO2A |
coronin, actin binding protein, 2A |
689 |
0.73 |
| chr16_19769260_19769411 | 0.15 |
CTD-2380F24.1 |
|
8031 |
0.18 |
| chr5_175968739_175969051 | 0.15 |
CDHR2 |
cadherin-related family member 2 |
617 |
0.6 |
| chr2_70154628_70154779 | 0.14 |
MXD1 |
MAX dimerization protein 1 |
12383 |
0.12 |
| chr7_151536159_151536670 | 0.14 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
24488 |
0.14 |
| chr21_15917916_15918619 | 0.14 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr16_31271694_31271922 | 0.14 |
ITGAM |
integrin, alpha M (complement component 3 receptor 3 subunit) |
493 |
0.67 |
| chr19_7894555_7894821 | 0.14 |
EVI5L |
ecotropic viral integration site 5-like |
431 |
0.73 |
| chr14_50053552_50053772 | 0.14 |
RN7SL1 |
RNA, 7SL, cytoplasmic 1 |
364 |
0.55 |
| chr17_79028934_79029085 | 0.14 |
BAIAP2 |
BAI1-associated protein 2 |
1153 |
0.39 |
| chr19_31472721_31472872 | 0.14 |
AC020952.1 |
Uncharacterized protein |
167566 |
0.04 |
| chr2_102973306_102973457 | 0.14 |
IL18R1 |
interleukin 18 receptor 1 |
992 |
0.54 |
| chr4_146018578_146019319 | 0.14 |
ANAPC10 |
anaphase promoting complex subunit 10 |
45 |
0.62 |
| chr7_32886100_32886251 | 0.14 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
44590 |
0.14 |
| chr1_92948652_92949059 | 0.14 |
GFI1 |
growth factor independent 1 transcription repressor |
656 |
0.79 |
| chr20_43810738_43810889 | 0.14 |
PI3 |
peptidase inhibitor 3, skin-derived |
7296 |
0.15 |
| chr2_218990770_218991030 | 0.14 |
CXCR2 |
chemokine (C-X-C motif) receptor 2 |
158 |
0.95 |
| chr2_162095067_162095304 | 0.14 |
TANK |
TRAF family member-associated NFKB activator |
7488 |
0.21 |
| chr15_75163179_75163478 | 0.14 |
SCAMP2 |
secretory carrier membrane protein 2 |
2053 |
0.19 |
| chr3_193560039_193560282 | 0.14 |
ENSG00000243991 |
. |
101650 |
0.07 |
| chr1_6054566_6054852 | 0.14 |
KCNAB2 |
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
1938 |
0.28 |
| chr9_132949942_132950122 | 0.14 |
NCS1 |
neuronal calcium sensor 1 |
12840 |
0.21 |
| chr1_182237824_182238059 | 0.14 |
ENSG00000206764 |
. |
58262 |
0.14 |
| chr1_234861269_234861851 | 0.14 |
ENSG00000201638 |
. |
112160 |
0.06 |
| chr11_111749891_111750174 | 0.14 |
C11orf1 |
chromosome 11 open reading frame 1 |
84 |
0.55 |
| chr1_28502388_28502717 | 0.14 |
PTAFR |
platelet-activating factor receptor |
1147 |
0.35 |
| chr19_54714818_54714969 | 0.14 |
RPS9 |
ribosomal protein S9 |
9865 |
0.07 |
| chr22_47338919_47339070 | 0.14 |
ENSG00000221672 |
. |
95191 |
0.08 |
| chr19_42052916_42053110 | 0.14 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
2873 |
0.23 |
| chr14_70161983_70162134 | 0.14 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
31559 |
0.19 |
| chr20_57225438_57225697 | 0.14 |
STX16 |
syntaxin 16 |
761 |
0.62 |
| chr1_161949318_161949469 | 0.14 |
OLFML2B |
olfactomedin-like 2B |
5629 |
0.27 |
| chr6_149639720_149640661 | 0.14 |
TAB2 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
754 |
0.68 |
| chr17_19015403_19015696 | 0.13 |
ENSG00000262202 |
. |
400 |
0.71 |
| chr6_26128396_26128716 | 0.13 |
HIST1H2AC |
histone cluster 1, H2ac |
4125 |
0.08 |
| chr2_166131422_166131573 | 0.13 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
19044 |
0.25 |
| chr16_11678677_11678828 | 0.13 |
LITAF |
lipopolysaccharide-induced TNF factor |
1477 |
0.42 |
| chr19_38826748_38826943 | 0.13 |
CATSPERG |
catsper channel auxiliary subunit gamma |
62 |
0.93 |
| chr8_21964305_21964456 | 0.13 |
NUDT18 |
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
2530 |
0.17 |
| chr8_142105442_142105593 | 0.13 |
DENND3 |
DENN/MADD domain containing 3 |
21860 |
0.19 |
| chr7_156797320_156797521 | 0.13 |
MNX1-AS2 |
MNX1 antisense RNA 2 |
1581 |
0.33 |
| chr12_53764513_53765147 | 0.13 |
SP1 |
Sp1 transcription factor |
9130 |
0.12 |
| chr19_41196777_41197167 | 0.13 |
NUMBL |
numb homolog (Drosophila)-like |
95 |
0.95 |
| chr16_30078571_30078900 | 0.13 |
ALDOA |
aldolase A, fructose-bisphosphate |
89 |
0.92 |
| chr11_119597594_119597745 | 0.13 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
1594 |
0.37 |
| chr14_65512386_65512664 | 0.13 |
ENSG00000266531 |
. |
1119 |
0.42 |
| chr15_40599321_40599893 | 0.13 |
PLCB2 |
phospholipase C, beta 2 |
419 |
0.68 |
| chr22_18250195_18250346 | 0.13 |
ENSG00000264757 |
. |
3245 |
0.23 |
| chr11_65342200_65342895 | 0.13 |
EHBP1L1 |
EH domain binding protein 1-like 1 |
962 |
0.29 |
| chr9_6703919_6704260 | 0.13 |
KDM4C |
lysine (K)-specific demethylase 4C |
16774 |
0.21 |
| chr19_41856873_41857286 | 0.13 |
TMEM91 |
transmembrane protein 91 |
263 |
0.82 |
| chr17_43306981_43307138 | 0.13 |
CTD-2020K17.1 |
|
7470 |
0.1 |
| chr8_97864758_97864909 | 0.13 |
CPQ |
carboxypeptidase Q |
91365 |
0.1 |
| chr15_65136767_65137537 | 0.13 |
AC069368.3 |
Uncharacterized protein |
3005 |
0.17 |
| chr3_11118976_11119127 | 0.13 |
SLC6A1-AS1 |
SLC6A1 antisense RNA 1 |
58141 |
0.12 |
| chr10_14611757_14612021 | 0.13 |
FAM107B |
family with sequence similarity 107, member B |
2140 |
0.39 |
| chr11_67186046_67186291 | 0.13 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
2484 |
0.1 |
| chr20_62670859_62671010 | 0.13 |
ZNF512B |
zinc finger protein 512B |
996 |
0.34 |
| chr4_81000146_81000297 | 0.13 |
ANTXR2 |
anthrax toxin receptor 2 |
5595 |
0.31 |
| chr15_40571810_40571961 | 0.13 |
ANKRD63 |
ankyrin repeat domain 63 |
2902 |
0.13 |
| chr3_65965301_65965465 | 0.13 |
ENSG00000202071 |
. |
24232 |
0.16 |
| chr17_39738951_39739102 | 0.13 |
KRT14 |
keratin 14 |
4147 |
0.12 |
| chr2_28691966_28692117 | 0.13 |
PLB1 |
phospholipase B1 |
12029 |
0.22 |
| chr15_69380999_69381150 | 0.13 |
RP11-809H16.4 |
|
45169 |
0.13 |
| chr1_210473057_210473208 | 0.12 |
HHAT |
hedgehog acyltransferase |
28464 |
0.19 |
| chr20_23067571_23067722 | 0.12 |
CD93 |
CD93 molecule |
669 |
0.69 |
| chr17_46607108_46607259 | 0.12 |
HOXB1 |
homeobox B1 |
1089 |
0.3 |
| chr20_20691484_20691635 | 0.12 |
RALGAPA2 |
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
1572 |
0.49 |
| chr10_105599968_105600119 | 0.12 |
SH3PXD2A |
SH3 and PX domains 2A |
15121 |
0.2 |
| chr6_157097564_157098248 | 0.12 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
1157 |
0.47 |
| chr8_19539783_19540152 | 0.12 |
CSGALNACT1 |
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
127 |
0.98 |
| chr19_11466484_11467017 | 0.12 |
DKFZP761J1410 |
Lipid phosphate phosphatase-related protein type 2 |
576 |
0.52 |
| chr6_12010262_12011396 | 0.12 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
1404 |
0.51 |
| chr20_10741569_10741720 | 0.12 |
JAG1 |
jagged 1 |
86950 |
0.09 |
| chr19_35494097_35494396 | 0.12 |
GRAMD1A |
GRAM domain containing 1A |
2886 |
0.14 |
| chr17_27223716_27224346 | 0.12 |
FLOT2 |
flotillin 2 |
639 |
0.35 |
| chr16_3057439_3057643 | 0.12 |
LA16c-380H5.2 |
|
2439 |
0.09 |
| chr12_72149024_72149971 | 0.12 |
RAB21 |
RAB21, member RAS oncogene family |
843 |
0.67 |
| chr16_85075183_85075477 | 0.12 |
KIAA0513 |
KIAA0513 |
13920 |
0.21 |
| chr12_14718728_14718879 | 0.12 |
RP11-695J4.2 |
|
1881 |
0.26 |
| chr8_121757630_121757781 | 0.12 |
RP11-713M15.1 |
|
15788 |
0.25 |
| chr16_85815842_85815993 | 0.12 |
ENSG00000252311 |
. |
412 |
0.72 |
| chr1_149608228_149608379 | 0.12 |
ENSG00000202496 |
. |
2386 |
0.21 |
| chr10_126697072_126697223 | 0.12 |
CTBP2 |
C-terminal binding protein 2 |
2565 |
0.29 |
| chr4_6912691_6912996 | 0.12 |
TBC1D14 |
TBC1 domain family, member 14 |
868 |
0.61 |
| chr18_34704825_34704976 | 0.12 |
KIAA1328 |
KIAA1328 |
58071 |
0.15 |
| chr3_175522251_175522402 | 0.12 |
NAALADL2-AS1 |
NAALADL2 antisense RNA 1 |
28205 |
0.2 |
| chr20_43150070_43150299 | 0.12 |
SERINC3 |
serine incorporator 3 |
467 |
0.76 |
| chr8_8945449_8945788 | 0.12 |
ENSG00000239078 |
. |
15592 |
0.15 |
| chr17_56595689_56596135 | 0.12 |
MTMR4 |
myotubularin related protein 4 |
646 |
0.52 |
| chr2_46561320_46561471 | 0.12 |
EPAS1 |
endothelial PAS domain protein 1 |
36854 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.1 | 0.2 | GO:0034113 | heterotypic cell-cell adhesion(GO:0034113) regulation of heterotypic cell-cell adhesion(GO:0034114) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.2 | GO:0032070 | regulation of deoxyribonuclease activity(GO:0032070) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.2 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.2 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.2 | GO:0061001 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.4 | GO:0035338 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.2 | GO:0045350 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.2 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.3 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.1 | GO:0097300 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.2 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0031440 | regulation of mRNA 3'-end processing(GO:0031440) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.1 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.0 | 0.0 | GO:0010979 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:1903224 | endodermal cell fate commitment(GO:0001711) endodermal cell fate specification(GO:0001714) endodermal cell differentiation(GO:0035987) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0032069 | regulation of nuclease activity(GO:0032069) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0043461 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.1 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0030201 | heparan sulfate proteoglycan metabolic process(GO:0030201) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.0 | GO:0002238 | response to molecule of fungal origin(GO:0002238) |
| 0.0 | 0.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:1902603 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.1 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.0 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.0 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.0 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:0002820 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.8 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:1902305 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.2 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.0 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.1 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0006623 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:1905145 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.0 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.0 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.0 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0060479 | lung cell differentiation(GO:0060479) lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.0 | GO:0045579 | positive regulation of B cell differentiation(GO:0045579) |
| 0.0 | 0.0 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.0 | GO:0010713 | negative regulation of collagen metabolic process(GO:0010713) negative regulation of multicellular organismal metabolic process(GO:0044252) |
| 0.0 | 0.3 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.4 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.1 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:1903115 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:0072179 | nephric duct formation(GO:0072179) |
| 0.0 | 0.0 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0045911 | positive regulation of DNA recombination(GO:0045911) |
| 0.0 | 0.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0052472 | modulation by host of viral transcription(GO:0043921) modulation by host of symbiont transcription(GO:0052472) |
| 0.0 | 0.0 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0015919 | peroxisomal membrane transport(GO:0015919) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0071732 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.0 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.0 | GO:0031094 | platelet dense tubular network(GO:0031094) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.0 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.0 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.2 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.2 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.1 | 0.3 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.3 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.0 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.0 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0015278 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0045569 | TRAIL binding(GO:0045569) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.1 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.0 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.1 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.0 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.0 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.0 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |