Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
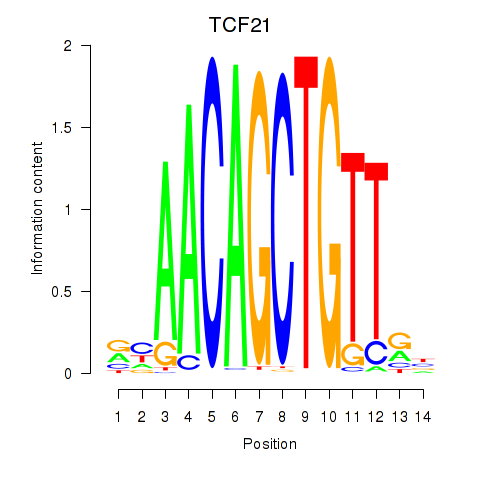
Results for TCF21
Z-value: 1.14
Transcription factors associated with TCF21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF21
|
ENSG00000118526.6 | transcription factor 21 |
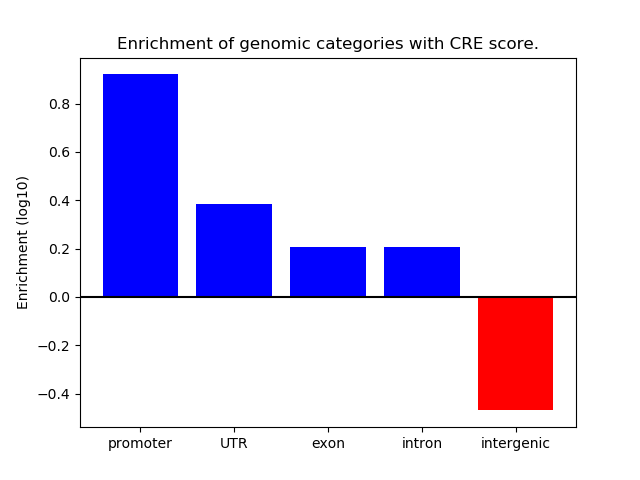
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_134210706_134210991 | TCF21 | 375 | 0.698482 | -0.31 | 4.2e-01 | Click! |
| chr6_134210535_134210686 | TCF21 | 137 | 0.830676 | -0.29 | 4.5e-01 | Click! |
| chr6_134211072_134211606 | TCF21 | 866 | 0.502462 | 0.21 | 5.8e-01 | Click! |
| chr6_134210328_134210483 | TCF21 | 68 | 0.757923 | 0.09 | 8.1e-01 | Click! |
Activity of the TCF21 motif across conditions
Conditions sorted by the z-value of the TCF21 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
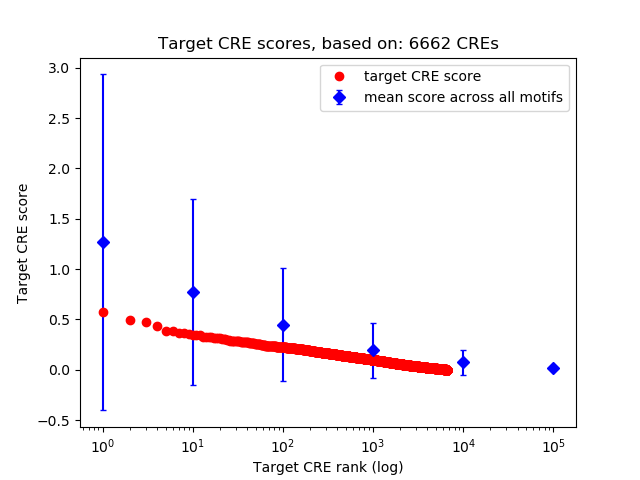
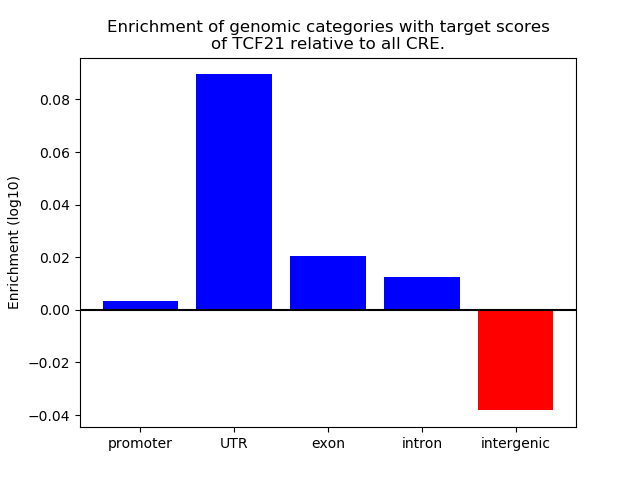
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_145276293_145276624 | 0.58 |
ZEB2-AS1 |
ZEB2 antisense RNA 1 |
434 |
0.52 |
| chr6_159465378_159465638 | 0.50 |
TAGAP |
T-cell activation RhoGTPase activating protein |
542 |
0.78 |
| chr20_31351724_31351875 | 0.47 |
DNMT3B |
DNA (cytosine-5-)-methyltransferase 3 beta |
1351 |
0.43 |
| chr19_15580169_15580596 | 0.44 |
RASAL3 |
RAS protein activator like 3 |
5000 |
0.14 |
| chr12_8862062_8862268 | 0.38 |
RIMKLB |
ribosomal modification protein rimK-like family member B |
9723 |
0.17 |
| chr20_52464106_52464266 | 0.38 |
AC005220.3 |
|
92513 |
0.08 |
| chr8_8610679_8610830 | 0.37 |
CLDN23 |
claudin 23 |
51306 |
0.15 |
| chr12_57828470_57828621 | 0.37 |
INHBC |
inhibin, beta C |
2 |
0.95 |
| chr15_57508769_57508920 | 0.35 |
TCF12 |
transcription factor 12 |
2784 |
0.37 |
| chr8_125617266_125617519 | 0.35 |
RP11-532M24.1 |
|
25520 |
0.16 |
| chr7_64349557_64349866 | 0.35 |
RP11-797H7.5 |
|
767 |
0.67 |
| chr12_9142516_9142764 | 0.34 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
423 |
0.81 |
| chr18_22811215_22811414 | 0.33 |
ZNF521 |
zinc finger protein 521 |
180 |
0.97 |
| chr14_75801139_75801383 | 0.33 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
54365 |
0.1 |
| chr1_245215310_245215673 | 0.33 |
ENSG00000251754 |
. |
8261 |
0.17 |
| chr19_16427219_16427370 | 0.32 |
CTD-2562J15.6 |
|
8031 |
0.16 |
| chr21_46970159_46970310 | 0.32 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
5909 |
0.24 |
| chr11_1874347_1875032 | 0.31 |
LSP1 |
lymphocyte-specific protein 1 |
489 |
0.65 |
| chr10_54514994_54515145 | 0.31 |
RP11-556E13.1 |
|
100 |
0.98 |
| chr1_114483508_114483659 | 0.31 |
HIPK1 |
homeodomain interacting protein kinase 1 |
10184 |
0.13 |
| chr4_82426507_82426753 | 0.31 |
RASGEF1B |
RasGEF domain family, member 1B |
33561 |
0.24 |
| chr8_41623788_41623939 | 0.30 |
ANK1 |
ankyrin 1, erythrocytic |
31277 |
0.15 |
| chr11_64512499_64513555 | 0.30 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
99 |
0.95 |
| chr3_184869954_184870148 | 0.29 |
C3orf70 |
chromosome 3 open reading frame 70 |
751 |
0.7 |
| chr17_68166046_68166268 | 0.29 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
481 |
0.55 |
| chr21_44524771_44524948 | 0.29 |
U2AF1 |
U2 small nuclear RNA auxiliary factor 1 |
503 |
0.75 |
| chr1_109204500_109204651 | 0.29 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
427 |
0.83 |
| chr17_143001_143152 | 0.29 |
RP11-1260E13.1 |
|
28107 |
0.13 |
| chr13_78493097_78493683 | 0.29 |
EDNRB |
endothelin receptor type B |
424 |
0.48 |
| chr1_245018870_245019021 | 0.29 |
HNRNPU-AS1 |
HNRNPU antisense RNA 1 |
146 |
0.95 |
| chr1_184121550_184121895 | 0.29 |
ENSG00000199840 |
. |
19235 |
0.25 |
| chr7_50357605_50357756 | 0.28 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
9362 |
0.29 |
| chr1_59782411_59782670 | 0.28 |
FGGY |
FGGY carbohydrate kinase domain containing |
6781 |
0.34 |
| chr11_67778123_67778316 | 0.28 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
429 |
0.73 |
| chr4_128553582_128553997 | 0.28 |
INTU |
inturned planar cell polarity protein |
298 |
0.94 |
| chr6_158622105_158622256 | 0.28 |
ENSG00000265803 |
. |
26164 |
0.16 |
| chr12_117036919_117037510 | 0.28 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
23558 |
0.24 |
| chr5_90478735_90478954 | 0.28 |
ENSG00000199643 |
. |
87700 |
0.1 |
| chr12_56138916_56139328 | 0.28 |
GDF11 |
growth differentiation factor 11 |
1939 |
0.16 |
| chr4_76822812_76822963 | 0.27 |
PPEF2 |
protein phosphatase, EF-hand calcium binding domain 2 |
794 |
0.58 |
| chr5_17144956_17145201 | 0.27 |
ENSG00000251990 |
. |
12102 |
0.2 |
| chr17_40283670_40283821 | 0.27 |
RAB5C |
RAB5C, member RAS oncogene family |
2968 |
0.13 |
| chr21_34443663_34444086 | 0.27 |
AP000282.2 |
|
464 |
0.55 |
| chr15_59978021_59978291 | 0.27 |
RP11-361D15.2 |
|
2560 |
0.21 |
| chr1_154171748_154172121 | 0.27 |
TPM3 |
tropomyosin 3 |
4810 |
0.1 |
| chr5_50007615_50007766 | 0.26 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
44299 |
0.21 |
| chr6_31555088_31555348 | 0.26 |
LST1 |
leukocyte specific transcript 1 |
125 |
0.86 |
| chr2_101841003_101841154 | 0.26 |
CNOT11 |
CCR4-NOT transcription complex, subunit 11 |
28186 |
0.16 |
| chr21_34185899_34186080 | 0.26 |
C21orf62 |
chromosome 21 open reading frame 62 |
0 |
0.97 |
| chr15_48909824_48909975 | 0.26 |
FBN1 |
fibrillin 1 |
28019 |
0.23 |
| chr7_148330575_148330726 | 0.26 |
C7orf33 |
chromosome 7 open reading frame 33 |
42993 |
0.14 |
| chr3_150995962_150996155 | 0.26 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
99 |
0.97 |
| chr14_91111245_91111396 | 0.26 |
TTC7B |
tetratricopeptide repeat domain 7B |
768 |
0.6 |
| chr4_153459378_153459529 | 0.26 |
ENSG00000268471 |
. |
1873 |
0.31 |
| chr9_71174970_71175121 | 0.25 |
TMEM252 |
transmembrane protein 252 |
19262 |
0.25 |
| chr6_133068019_133068170 | 0.25 |
RP1-55C23.7 |
|
5720 |
0.13 |
| chr9_6913439_6913590 | 0.25 |
RP11-403H13.1 |
|
10844 |
0.24 |
| chr14_23288815_23289060 | 0.25 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
7 |
0.95 |
| chr6_136791083_136791234 | 0.25 |
MAP7 |
microtubule-associated protein 7 |
3145 |
0.32 |
| chr6_46376004_46376155 | 0.25 |
RCAN2 |
regulator of calcineurin 2 |
48634 |
0.16 |
| chr20_52703439_52703654 | 0.24 |
BCAS1 |
breast carcinoma amplified sequence 1 |
16242 |
0.21 |
| chr1_160681036_160681588 | 0.24 |
CD48 |
CD48 molecule |
281 |
0.89 |
| chr11_32460808_32460959 | 0.24 |
WT1-AS |
WT1 antisense RNA |
967 |
0.61 |
| chr4_15418160_15418311 | 0.24 |
RP11-484O2.1 |
|
3421 |
0.22 |
| chrX_131624482_131624759 | 0.24 |
MBNL3 |
muscleblind-like splicing regulator 3 |
624 |
0.83 |
| chr12_14796282_14796433 | 0.24 |
RP11-174G6.1 |
|
22232 |
0.16 |
| chr2_115918703_115918854 | 0.24 |
DPP10-AS1 |
DPP10 antisense RNA 1 |
10 |
0.88 |
| chr12_123752691_123752842 | 0.24 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
65 |
0.95 |
| chr17_61778306_61778706 | 0.24 |
LIMD2 |
LIM domain containing 2 |
26 |
0.97 |
| chr11_8228040_8228278 | 0.24 |
RIC3 |
RIC3 acetylcholine receptor chaperone |
37557 |
0.16 |
| chr7_55601634_55601785 | 0.24 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
3678 |
0.29 |
| chr5_123392522_123392673 | 0.24 |
CSNK1G3 |
casein kinase 1, gamma 3 |
468478 |
0.01 |
| chr3_23804583_23804865 | 0.24 |
ENSG00000238672 |
. |
4378 |
0.25 |
| chr1_108593520_108593821 | 0.24 |
ENSG00000264753 |
. |
32277 |
0.19 |
| chr7_88025228_88025379 | 0.24 |
ENSG00000207094 |
. |
53243 |
0.15 |
| chr10_92690511_92690662 | 0.24 |
ENSG00000201604 |
. |
7994 |
0.19 |
| chr15_86126252_86126683 | 0.23 |
RP11-815J21.2 |
|
3058 |
0.23 |
| chr1_211500199_211501024 | 0.23 |
TRAF5 |
TNF receptor-associated factor 5 |
432 |
0.87 |
| chr1_234910955_234911218 | 0.23 |
ENSG00000201638 |
. |
62634 |
0.13 |
| chr11_128588908_128589084 | 0.23 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23078 |
0.17 |
| chr9_13787686_13787970 | 0.23 |
NFIB |
nuclear factor I/B |
392963 |
0.01 |
| chr2_127417167_127417318 | 0.23 |
GYPC |
glycophorin C (Gerbich blood group) |
3482 |
0.34 |
| chr2_168438647_168438800 | 0.23 |
ENSG00000238357 |
. |
49328 |
0.19 |
| chr5_35781420_35781571 | 0.23 |
SPEF2 |
sperm flagellar 2 |
2225 |
0.36 |
| chr17_66596046_66596347 | 0.23 |
FAM20A |
family with sequence similarity 20, member A |
1334 |
0.52 |
| chr6_24919733_24919934 | 0.23 |
FAM65B |
family with sequence similarity 65, member B |
8638 |
0.23 |
| chr12_75875877_75876340 | 0.23 |
GLIPR1 |
GLI pathogenesis-related 1 |
1124 |
0.49 |
| chr2_174890200_174890425 | 0.23 |
SP3 |
Sp3 transcription factor |
59882 |
0.15 |
| chr11_35144138_35144289 | 0.23 |
AL356215.1 |
|
6002 |
0.19 |
| chr15_63524002_63524153 | 0.23 |
RAB8B |
RAB8B, member RAS oncogene family |
42272 |
0.14 |
| chr5_137691113_137691264 | 0.23 |
KDM3B |
lysine (K)-specific demethylase 3B |
2903 |
0.18 |
| chr3_107821041_107821248 | 0.23 |
CD47 |
CD47 molecule |
11272 |
0.29 |
| chr17_3865236_3865510 | 0.23 |
ATP2A3 |
ATPase, Ca++ transporting, ubiquitous |
2212 |
0.29 |
| chr10_112357348_112357499 | 0.23 |
ENSG00000239125 |
. |
21305 |
0.14 |
| chr11_123229225_123229659 | 0.23 |
ENSG00000265357 |
. |
22778 |
0.2 |
| chr2_43455773_43455924 | 0.23 |
AC010883.5 |
|
864 |
0.61 |
| chr5_95066383_95066820 | 0.23 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
25 |
0.96 |
| chr11_118110290_118110538 | 0.23 |
MPZL3 |
myelin protein zero-like 3 |
12648 |
0.13 |
| chr13_24210772_24210935 | 0.23 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
57334 |
0.14 |
| chr1_31460456_31460607 | 0.23 |
PUM1 |
pumilio RNA-binding family member 1 |
7375 |
0.18 |
| chr16_23161362_23161529 | 0.22 |
USP31 |
ubiquitin specific peptidase 31 |
854 |
0.66 |
| chr12_121677598_121677871 | 0.22 |
CAMKK2 |
calcium/calmodulin-dependent protein kinase kinase 2, beta |
20611 |
0.16 |
| chr1_110159560_110159711 | 0.22 |
AMPD2 |
adenosine monophosphate deaminase 2 |
909 |
0.36 |
| chr1_151592946_151593154 | 0.22 |
RP11-404E16.1 |
|
7211 |
0.11 |
| chr17_38604215_38604366 | 0.22 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
4577 |
0.16 |
| chr19_12889048_12889583 | 0.22 |
HOOK2 |
hook microtubule-tethering protein 2 |
44 |
0.92 |
| chr2_74804644_74804826 | 0.22 |
LOXL3 |
lysyl oxidase-like 3 |
21918 |
0.09 |
| chr2_60638627_60638831 | 0.22 |
ENSG00000266078 |
. |
24149 |
0.2 |
| chr11_118085317_118085565 | 0.22 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
165 |
0.94 |
| chrX_128914480_128914899 | 0.22 |
SASH3 |
SAM and SH3 domain containing 3 |
729 |
0.68 |
| chr12_111618967_111619231 | 0.22 |
CUX2 |
cut-like homeobox 2 |
81849 |
0.09 |
| chr9_93564472_93565060 | 0.22 |
SYK |
spleen tyrosine kinase |
557 |
0.87 |
| chr3_105295301_105295452 | 0.22 |
ALCAM |
activated leukocyte cell adhesion molecule |
42930 |
0.21 |
| chr10_130832704_130832855 | 0.22 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
432669 |
0.01 |
| chr1_212325647_212325798 | 0.22 |
ENSG00000252879 |
. |
52704 |
0.12 |
| chr18_56245541_56245920 | 0.22 |
RP11-126O1.2 |
|
21428 |
0.14 |
| chr14_35179286_35179550 | 0.22 |
CFL2 |
cofilin 2 (muscle) |
3616 |
0.25 |
| chr14_88474301_88474452 | 0.22 |
GPR65 |
G protein-coupled receptor 65 |
2908 |
0.23 |
| chr5_141872256_141872598 | 0.22 |
ENSG00000252831 |
. |
41483 |
0.18 |
| chr8_120650260_120650433 | 0.22 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
674 |
0.78 |
| chr11_122536513_122536664 | 0.21 |
UBASH3B |
ubiquitin associated and SH3 domain containing B |
10205 |
0.24 |
| chr2_238598166_238598563 | 0.21 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
2424 |
0.34 |
| chr6_32920569_32920784 | 0.21 |
HLA-DMA |
major histocompatibility complex, class II, DM alpha |
162 |
0.56 |
| chr2_40907638_40908036 | 0.21 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
69644 |
0.14 |
| chr11_42107655_42107806 | 0.21 |
LRRC4C |
leucine rich repeat containing 4C |
626407 |
0.0 |
| chr10_94515647_94515798 | 0.21 |
ENSG00000201412 |
. |
45357 |
0.14 |
| chr18_66505598_66505749 | 0.21 |
CCDC102B |
coiled-coil domain containing 102B |
1672 |
0.41 |
| chr8_116675050_116675578 | 0.21 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr15_69695885_69696036 | 0.21 |
ENSG00000207395 |
. |
2820 |
0.17 |
| chr4_87050749_87050921 | 0.21 |
RP11-778J15.1 |
|
9968 |
0.23 |
| chr8_74283083_74283286 | 0.21 |
RP11-434I12.2 |
|
14488 |
0.25 |
| chr9_139423244_139423395 | 0.21 |
ENSG00000263403 |
. |
9241 |
0.09 |
| chr18_9083293_9083444 | 0.21 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
19260 |
0.17 |
| chr8_117163981_117164132 | 0.21 |
ENSG00000221793 |
. |
15058 |
0.29 |
| chr15_63524568_63524719 | 0.21 |
RAB8B |
RAB8B, member RAS oncogene family |
42838 |
0.14 |
| chr13_103423092_103423243 | 0.21 |
TEX30 |
testis expressed 30 |
17 |
0.96 |
| chr2_74264500_74264778 | 0.21 |
TET3 |
tet methylcytosine dioxygenase 3 |
8811 |
0.18 |
| chr11_63319075_63319226 | 0.21 |
HRASLS2 |
HRAS-like suppressor 2 |
11705 |
0.14 |
| chr15_63795725_63795911 | 0.21 |
USP3 |
ubiquitin specific peptidase 3 |
975 |
0.63 |
| chr10_26778557_26778708 | 0.21 |
ENSG00000199733 |
. |
19886 |
0.24 |
| chr2_233949302_233949453 | 0.21 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
24188 |
0.17 |
| chr9_83728640_83728791 | 0.21 |
ENSG00000221581 |
. |
379471 |
0.01 |
| chr18_52519559_52519845 | 0.21 |
RAB27B |
RAB27B, member RAS oncogene family |
24272 |
0.21 |
| chr13_77899686_77900769 | 0.21 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
587 |
0.85 |
| chr18_52495652_52496658 | 0.21 |
RAB27B |
RAB27B, member RAS oncogene family |
725 |
0.77 |
| chr2_118773318_118773469 | 0.21 |
ENSG00000243510 |
. |
565 |
0.64 |
| chr13_99195550_99195827 | 0.21 |
STK24 |
serine/threonine kinase 24 |
21436 |
0.2 |
| chr7_138293225_138293376 | 0.21 |
ENSG00000252188 |
. |
16624 |
0.23 |
| chr21_46573190_46573341 | 0.21 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
18292 |
0.17 |
| chr8_1904836_1904987 | 0.21 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
17133 |
0.24 |
| chr4_53017131_53017386 | 0.21 |
SPATA18 |
spermatogenesis associated 18 |
99676 |
0.08 |
| chr3_133661654_133661805 | 0.21 |
C3orf36 |
chromosome 3 open reading frame 36 |
13073 |
0.23 |
| chr8_23997445_23997596 | 0.20 |
ENSG00000207201 |
. |
62849 |
0.14 |
| chr17_45334525_45334676 | 0.20 |
ENSG00000238419 |
. |
2186 |
0.21 |
| chr6_13273500_13273733 | 0.20 |
PHACTR1 |
phosphatase and actin regulator 1 |
627 |
0.46 |
| chr6_107021251_107021402 | 0.20 |
AIM1 |
absent in melanoma 1 |
32297 |
0.14 |
| chr19_42260620_42260771 | 0.20 |
CEACAM6 |
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
1366 |
0.33 |
| chr9_134150260_134150411 | 0.20 |
FAM78A |
family with sequence similarity 78, member A |
1599 |
0.36 |
| chr21_36612358_36612509 | 0.20 |
RUNX1 |
runt-related transcription factor 1 |
190792 |
0.03 |
| chr8_23366659_23366810 | 0.20 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
19584 |
0.16 |
| chr3_9004693_9005124 | 0.20 |
RAD18 |
RAD18 homolog (S. cerevisiae) |
278 |
0.92 |
| chr1_167619596_167619862 | 0.20 |
RP3-455J7.4 |
|
19818 |
0.18 |
| chr3_151963057_151963208 | 0.20 |
MBNL1 |
muscleblind-like splicing regulator 1 |
22697 |
0.21 |
| chr3_15313694_15313943 | 0.20 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
3598 |
0.18 |
| chr4_120291951_120292186 | 0.20 |
ENSG00000201186 |
. |
3351 |
0.26 |
| chr18_9113256_9113407 | 0.20 |
RP11-143J12.3 |
|
2544 |
0.24 |
| chr8_121822100_121822251 | 0.20 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
2208 |
0.3 |
| chr6_110555384_110555535 | 0.20 |
CDC40 |
cell division cycle 40 |
53835 |
0.13 |
| chr3_18487175_18487565 | 0.20 |
SATB1 |
SATB homeobox 1 |
290 |
0.75 |
| chr2_233948085_233948236 | 0.20 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
22971 |
0.18 |
| chr2_172877202_172877353 | 0.20 |
METAP1D |
methionyl aminopeptidase type 1D (mitochondrial) |
12787 |
0.23 |
| chr1_8761101_8761375 | 0.20 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
2040 |
0.41 |
| chr6_75877058_75877209 | 0.20 |
COL12A1 |
collagen, type XII, alpha 1 |
15228 |
0.25 |
| chr12_76413840_76414001 | 0.20 |
RP11-290L1.3 |
|
10354 |
0.19 |
| chr1_246945751_246945958 | 0.20 |
ENSG00000227953 |
. |
6460 |
0.17 |
| chrX_118407364_118407689 | 0.20 |
PGRMC1 |
progesterone receptor membrane component 1 |
37238 |
0.17 |
| chr9_6959941_6960092 | 0.20 |
KDM4C |
lysine (K)-specific demethylase 4C |
34515 |
0.21 |
| chr7_131240036_131240570 | 0.20 |
PODXL |
podocalyxin-like |
1063 |
0.65 |
| chr8_97523748_97523899 | 0.20 |
SDC2 |
syndecan 2 |
17587 |
0.25 |
| chr17_48218383_48218580 | 0.20 |
AC002401.1 |
|
5530 |
0.1 |
| chr13_33752733_33752884 | 0.20 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
7408 |
0.23 |
| chr5_142381326_142381532 | 0.20 |
ARHGAP26 |
Rho GTPase activating protein 26 |
35332 |
0.22 |
| chr9_81972295_81972446 | 0.20 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
214318 |
0.03 |
| chr1_150972321_150972472 | 0.20 |
FAM63A |
family with sequence similarity 63, member A |
1750 |
0.17 |
| chr3_184893391_184893626 | 0.20 |
EHHADH-AS1 |
EHHADH antisense RNA 1 |
12819 |
0.2 |
| chr4_65504939_65505090 | 0.19 |
TECRL |
trans-2,3-enoyl-CoA reductase-like |
229828 |
0.02 |
| chr4_6927460_6927611 | 0.19 |
TBC1D14 |
TBC1 domain family, member 14 |
15560 |
0.16 |
| chr5_156540231_156540382 | 0.19 |
HAVCR2 |
hepatitis A virus cellular receptor 2 |
3581 |
0.17 |
| chr1_66868301_66868452 | 0.19 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
48311 |
0.18 |
| chr18_53254177_53254364 | 0.19 |
TCF4 |
transcription factor 4 |
77 |
0.98 |
| chr2_207938706_207938857 | 0.19 |
ENSG00000253008 |
. |
36016 |
0.16 |
| chr6_112960793_112960944 | 0.19 |
ENSG00000252215 |
. |
107484 |
0.07 |
| chr13_114064294_114064563 | 0.19 |
ADPRHL1 |
ADP-ribosylhydrolase like 1 |
39029 |
0.11 |
| chr2_101478838_101478989 | 0.19 |
AC092168.2 |
|
39790 |
0.16 |
| chr10_88170804_88170968 | 0.19 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
44651 |
0.14 |
| chr12_123713655_123713840 | 0.19 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
1246 |
0.35 |
| chr20_10457057_10457208 | 0.19 |
SLX4IP |
SLX4 interacting protein |
41181 |
0.16 |
| chr18_60605343_60605494 | 0.19 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
35035 |
0.2 |
| chr1_162469519_162469670 | 0.19 |
UHMK1 |
U2AF homology motif (UHM) kinase 1 |
1961 |
0.33 |
| chr12_10876110_10876483 | 0.19 |
YBX3 |
Y box binding protein 3 |
385 |
0.85 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.1 | 0.2 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.1 | 0.2 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.2 | GO:0014041 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.1 | 0.2 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.3 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.3 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0032661 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.2 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.2 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0070339 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.2 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0001714 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.2 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.0 | GO:1903670 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0002068 | glandular epithelial cell development(GO:0002068) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0015811 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.0 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.0 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.0 | GO:0002246 | wound healing involved in inflammatory response(GO:0002246) connective tissue replacement involved in inflammatory response wound healing(GO:0002248) inflammatory response to wounding(GO:0090594) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0010829 | negative regulation of glucose transport(GO:0010829) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.0 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.0 | GO:0045354 | interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.0 | GO:0042819 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.0 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.0 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.0 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.0 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.0 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0032691 | negative regulation of interleukin-1 beta production(GO:0032691) negative regulation of interleukin-1 production(GO:0032692) |
| 0.0 | 0.0 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0006534 | cysteine metabolic process(GO:0006534) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0043388 | positive regulation of DNA binding(GO:0043388) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.1 | GO:0006853 | carnitine shuttle(GO:0006853) carnitine transmembrane transport(GO:1902603) |
| 0.0 | 0.1 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.0 | GO:0052167 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.0 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.0 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.0 | 0.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.0 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.0 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0030312 | external encapsulating structure(GO:0030312) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.3 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.0 | GO:0033185 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.0 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.0 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.3 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.2 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.0 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.0 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.0 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0050542 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.5 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID S1P S1P3 PATHWAY | S1P3 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME PI3K CASCADE | Genes involved in PI3K Cascade |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.7 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.0 | REACTOME PLATELET HOMEOSTASIS | Genes involved in Platelet homeostasis |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.0 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |