Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for TCF3_MYOG
Z-value: 0.35
Transcription factors associated with TCF3_MYOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
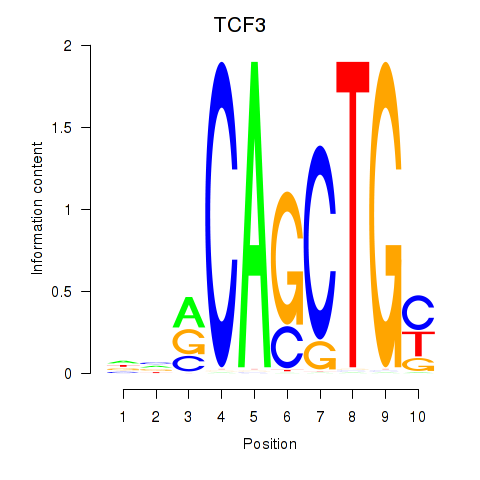
TCF3
|
ENSG00000071564.10 | transcription factor 3 |
|
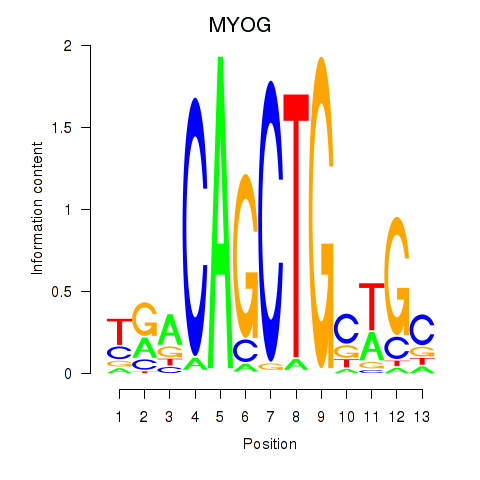
MYOG
|
ENSG00000122180.4 | myogenin |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_203044604_203044831 | MYOG | 10447 | 0.130208 | -0.49 | 1.8e-01 | Click! |
| chr1_203052021_203052172 | MYOG | 3068 | 0.179211 | -0.35 | 3.5e-01 | Click! |
| chr1_203044981_203045143 | MYOG | 10102 | 0.130813 | -0.21 | 5.9e-01 | Click! |
| chr1_203045268_203045454 | MYOG | 9803 | 0.131356 | -0.06 | 8.7e-01 | Click! |
| chr19_1682723_1682874 | TCF3 | 30194 | 0.105053 | -0.62 | 7.7e-02 | Click! |
| chr19_1675853_1676228 | TCF3 | 23436 | 0.114431 | -0.57 | 1.1e-01 | Click! |
| chr19_1653201_1653411 | TCF3 | 702 | 0.573603 | 0.42 | 2.6e-01 | Click! |
| chr19_1701461_1701612 | TCF3 | 48932 | 0.076519 | -0.41 | 2.7e-01 | Click! |
| chr19_1650811_1650962 | TCF3 | 142 | 0.935336 | -0.39 | 3.0e-01 | Click! |
Activity of the TCF3_MYOG motif across conditions
Conditions sorted by the z-value of the TCF3_MYOG motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
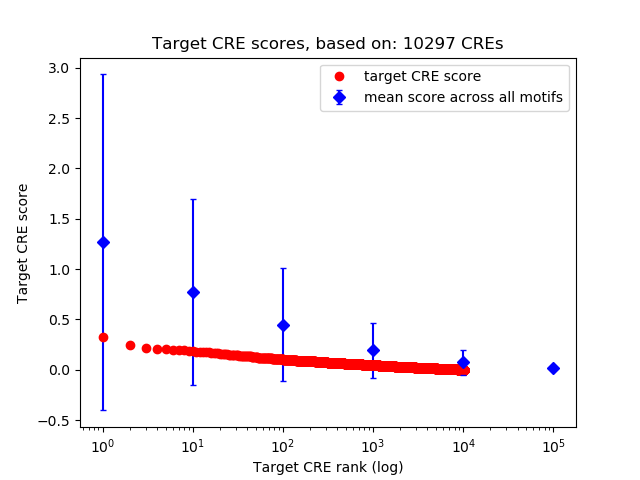
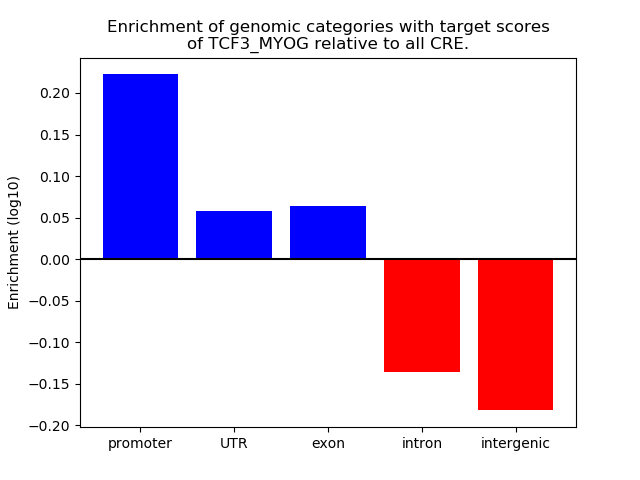
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_160681036_160681588 | 0.33 |
CD48 |
CD48 molecule |
281 |
0.89 |
| chr22_42177525_42177912 | 0.25 |
MEI1 |
meiosis inhibitor 1 |
21 |
0.96 |
| chr22_21738155_21738638 | 0.22 |
RIMBP3B |
RIMS binding protein 3B |
733 |
0.51 |
| chr22_20460794_20461266 | 0.21 |
RIMBP3 |
RIMS binding protein 3 |
756 |
0.52 |
| chr11_84028149_84028546 | 0.20 |
DLG2 |
discs, large homolog 2 (Drosophila) |
35 |
0.99 |
| chr12_133006842_133006993 | 0.20 |
MUC8 |
mucin 8 |
43809 |
0.14 |
| chrX_13103844_13104411 | 0.20 |
FAM9C |
family with sequence similarity 9, member C |
41326 |
0.19 |
| chr2_240188602_240189192 | 0.19 |
ENSG00000265215 |
. |
38260 |
0.14 |
| chr22_21904755_21905186 | 0.19 |
RIMBP3C |
RIMS binding protein 3C |
150 |
0.88 |
| chr15_60419498_60420019 | 0.18 |
FOXB1 |
forkhead box B1 |
123337 |
0.06 |
| chr2_25562492_25562759 | 0.18 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
2129 |
0.34 |
| chr5_169729652_169729829 | 0.18 |
LCP2 |
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) |
4509 |
0.25 |
| chr5_150535420_150535844 | 0.18 |
ANXA6 |
annexin A6 |
1676 |
0.39 |
| chr19_3180678_3181240 | 0.17 |
S1PR4 |
sphingosine-1-phosphate receptor 4 |
2223 |
0.2 |
| chr11_118212224_118212431 | 0.17 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
675 |
0.54 |
| chr1_65530319_65530470 | 0.17 |
ENSG00000199135 |
. |
6203 |
0.2 |
| chr11_65407442_65407916 | 0.17 |
SIPA1 |
signal-induced proliferation-associated 1 |
87 |
0.92 |
| chr20_56195172_56195527 | 0.16 |
ZBP1 |
Z-DNA binding protein 1 |
101 |
0.98 |
| chr13_52398191_52398536 | 0.16 |
RP11-327P2.5 |
|
19930 |
0.17 |
| chr10_73531816_73531986 | 0.16 |
C10orf54 |
chromosome 10 open reading frame 54 |
1354 |
0.44 |
| chr10_104420612_104420763 | 0.16 |
TRIM8 |
tripartite motif containing 8 |
16043 |
0.16 |
| chr18_2967390_2967541 | 0.16 |
RP11-737O24.1 |
|
449 |
0.78 |
| chr3_50646848_50647098 | 0.16 |
CISH |
cytokine inducible SH2-containing protein |
2230 |
0.22 |
| chr19_48835948_48836196 | 0.15 |
EMP3 |
epithelial membrane protein 3 |
7207 |
0.11 |
| chr13_75900551_75900966 | 0.15 |
TBC1D4 |
TBC1 domain family, member 4 |
14909 |
0.24 |
| chr18_43266884_43267229 | 0.15 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
20949 |
0.17 |
| chr3_114832402_114832553 | 0.15 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
33641 |
0.25 |
| chrX_44203680_44203969 | 0.15 |
EFHC2 |
EF-hand domain (C-terminal) containing 2 |
906 |
0.71 |
| chr20_35274684_35274835 | 0.15 |
SLA2 |
Src-like-adaptor 2 |
140 |
0.94 |
| chr2_220408420_220408618 | 0.14 |
CHPF |
chondroitin polymerizing factor |
10 |
0.62 |
| chr14_91840122_91840324 | 0.14 |
ENSG00000265856 |
. |
40166 |
0.15 |
| chrY_15863423_15864046 | 0.14 |
KALP |
Kallmann syndrome sequence pseudogene |
198 |
0.96 |
| chr8_128988445_128988635 | 0.14 |
ENSG00000221771 |
. |
15661 |
0.16 |
| chr7_150434707_150435064 | 0.14 |
GIMAP5 |
GTPase, IMAP family member 5 |
449 |
0.8 |
| chr6_144655565_144656031 | 0.14 |
RP1-91J24.3 |
|
1361 |
0.51 |
| chr1_244215459_244215610 | 0.14 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
949 |
0.61 |
| chr7_72300733_72300925 | 0.14 |
SBDSP1 |
Shwachman-Bodian-Diamond syndrome pseudogene 1 |
594 |
0.6 |
| chr11_2323777_2324127 | 0.14 |
TSPAN32 |
tetraspanin 32 |
1 |
0.92 |
| chr7_150148052_150148382 | 0.14 |
GIMAP8 |
GTPase, IMAP family member 8 |
499 |
0.78 |
| chr17_4615541_4615774 | 0.14 |
ARRB2 |
arrestin, beta 2 |
1663 |
0.18 |
| chr20_50158140_50158637 | 0.14 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
870 |
0.72 |
| chr22_24039509_24039689 | 0.14 |
RGL4 |
ral guanine nucleotide dissociation stimulator-like 4 |
928 |
0.45 |
| chr6_32813075_32813316 | 0.13 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
582 |
0.44 |
| chr16_27413508_27414237 | 0.13 |
IL21R |
interleukin 21 receptor |
377 |
0.88 |
| chr4_16219327_16219478 | 0.13 |
TAPT1 |
transmembrane anterior posterior transformation 1 |
8692 |
0.2 |
| chr14_106854492_106854843 | 0.13 |
IGHV3-37 |
immunoglobulin heavy variable 3-37 (pseudogene) |
1641 |
0.1 |
| chr17_80260072_80260536 | 0.13 |
CD7 |
CD7 molecule |
15124 |
0.1 |
| chr16_28996496_28996848 | 0.13 |
LAT |
linker for activation of T cells |
11 |
0.94 |
| chr15_21995486_21996039 | 0.13 |
DKFZP547L112 |
|
15608 |
0.19 |
| chr15_75082884_75083035 | 0.13 |
ENSG00000264386 |
. |
1861 |
0.2 |
| chr10_129985976_129986188 | 0.13 |
MKI67 |
marker of proliferation Ki-67 |
61433 |
0.15 |
| chr1_209930383_209930698 | 0.12 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
1016 |
0.45 |
| chrX_153267742_153268222 | 0.12 |
IRAK1 |
interleukin-1 receptor-associated kinase 1 |
11715 |
0.09 |
| chr15_29211815_29211966 | 0.12 |
APBA2 |
amyloid beta (A4) precursor protein-binding, family A, member 2 |
320 |
0.92 |
| chr1_225654970_225655492 | 0.12 |
RP11-496N12.6 |
|
2186 |
0.34 |
| chr22_32598633_32598888 | 0.12 |
RFPL2 |
ret finger protein-like 2 |
704 |
0.6 |
| chr17_43305439_43305590 | 0.12 |
CTD-2020K17.1 |
|
5925 |
0.1 |
| chr11_1874347_1875032 | 0.12 |
LSP1 |
lymphocyte-specific protein 1 |
489 |
0.65 |
| chr2_225803456_225803653 | 0.12 |
DOCK10 |
dedicator of cytokinesis 10 |
8228 |
0.3 |
| chr15_20988807_20989390 | 0.12 |
AC012414.1 |
Uncharacterized protein |
15589 |
0.17 |
| chr17_8868395_8868683 | 0.12 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
485 |
0.85 |
| chr11_67186046_67186291 | 0.12 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
2484 |
0.1 |
| chr22_22292604_22292980 | 0.12 |
LL22NC03-86G7.1 |
|
127 |
0.51 |
| chr17_38720511_38720770 | 0.12 |
CCR7 |
chemokine (C-C motif) receptor 7 |
1071 |
0.49 |
| chr20_62369505_62369786 | 0.12 |
RP4-583P15.14 |
|
22 |
0.92 |
| chr11_67176924_67177082 | 0.12 |
TBC1D10C |
TBC1 domain family, member 10C |
5343 |
0.07 |
| chr6_31238477_31238986 | 0.12 |
HLA-C |
major histocompatibility complex, class I, C |
1115 |
0.41 |
| chr17_4617667_4617818 | 0.12 |
ARRB2 |
arrestin, beta 2 |
1142 |
0.27 |
| chr1_169677205_169677356 | 0.11 |
SELL |
selectin L |
3559 |
0.24 |
| chr1_145435410_145435648 | 0.11 |
TXNIP |
thioredoxin interacting protein |
2940 |
0.16 |
| chr20_62270981_62271132 | 0.11 |
CTD-3184A7.4 |
|
12453 |
0.09 |
| chr8_65607910_65608061 | 0.11 |
RP11-1D12.1 |
|
39031 |
0.2 |
| chr1_154315658_154315860 | 0.11 |
ENSG00000238365 |
. |
4540 |
0.12 |
| chr2_68994932_68995083 | 0.11 |
ARHGAP25 |
Rho GTPase activating protein 25 |
6926 |
0.26 |
| chr17_75456326_75456913 | 0.11 |
SEPT9 |
septin 9 |
4171 |
0.18 |
| chr10_6620632_6620783 | 0.11 |
PRKCQ |
protein kinase C, theta |
1494 |
0.57 |
| chr2_10522774_10523051 | 0.11 |
HPCAL1 |
hippocalcin-like 1 |
37235 |
0.15 |
| chr13_31310422_31310683 | 0.11 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
907 |
0.71 |
| chr17_33568427_33568649 | 0.11 |
RP11-799D4.4 |
|
1444 |
0.28 |
| chr17_1478951_1479102 | 0.11 |
PITPNA |
phosphatidylinositol transfer protein, alpha |
12916 |
0.11 |
| chr21_35322492_35322643 | 0.11 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
1337 |
0.41 |
| chr22_50523207_50523373 | 0.11 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
398 |
0.79 |
| chr19_5132131_5132282 | 0.11 |
CTC-482H14.5 |
|
45924 |
0.14 |
| chr14_77013139_77013290 | 0.11 |
ESRRB |
estrogen-related receptor beta |
139333 |
0.04 |
| chrX_70331169_70331370 | 0.11 |
IL2RG |
interleukin 2 receptor, gamma |
29 |
0.95 |
| chr7_127749300_127749451 | 0.11 |
ENSG00000207588 |
. |
27462 |
0.2 |
| chr2_113932645_113932882 | 0.11 |
AC016683.5 |
|
151 |
0.91 |
| chr18_5294330_5295063 | 0.11 |
ZBTB14 |
zinc finger and BTB domain containing 14 |
127 |
0.98 |
| chr6_28108040_28108408 | 0.11 |
ZKSCAN8 |
zinc finger with KRAB and SCAN domains 8 |
1492 |
0.26 |
| chr12_112449785_112449936 | 0.11 |
TMEM116 |
transmembrane protein 116 |
1055 |
0.37 |
| chr7_55636821_55636978 | 0.11 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
2662 |
0.29 |
| chr7_43690953_43691104 | 0.11 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
1133 |
0.55 |
| chr6_11786252_11786455 | 0.10 |
ADTRP |
androgen-dependent TFPI-regulating protein |
6492 |
0.3 |
| chrX_21391687_21391849 | 0.10 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
768 |
0.79 |
| chr2_197034753_197035766 | 0.10 |
STK17B |
serine/threonine kinase 17b |
465 |
0.83 |
| chr10_105670056_105670248 | 0.10 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
7275 |
0.18 |
| chr9_117149358_117150269 | 0.10 |
AKNA |
AT-hook transcription factor |
430 |
0.85 |
| chr12_54892329_54892523 | 0.10 |
NCKAP1L |
NCK-associated protein 1-like |
142 |
0.94 |
| chr7_66459685_66459882 | 0.10 |
SBDS |
Shwachman-Bodian-Diamond syndrome |
805 |
0.55 |
| chr12_109027255_109027564 | 0.10 |
SELPLG |
selectin P ligand |
261 |
0.87 |
| chr5_176560848_176561583 | 0.10 |
NSD1 |
nuclear receptor binding SET domain protein 1 |
15 |
0.98 |
| chr17_76117639_76118201 | 0.10 |
TMC6 |
transmembrane channel-like 6 |
909 |
0.44 |
| chr10_30994119_30994515 | 0.10 |
SVILP1 |
supervillin pseudogene 1 |
9467 |
0.26 |
| chr1_154924890_154925041 | 0.10 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
3615 |
0.1 |
| chr7_152373399_152373635 | 0.10 |
XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
267 |
0.92 |
| chr6_91005254_91005982 | 0.10 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
843 |
0.69 |
| chr6_42408755_42408959 | 0.10 |
TRERF1 |
transcriptional regulating factor 1 |
10142 |
0.2 |
| chr14_90863482_90863819 | 0.10 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
250 |
0.92 |
| chr14_22471633_22471925 | 0.10 |
ENSG00000238634 |
. |
139108 |
0.04 |
| chr1_2221478_2221737 | 0.10 |
RP4-713A8.1 |
|
36525 |
0.09 |
| chr17_75125901_75126097 | 0.10 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
2044 |
0.33 |
| chr6_159235122_159235362 | 0.10 |
EZR-AS1 |
EZR antisense RNA 1 |
3801 |
0.21 |
| chr6_2901280_2901469 | 0.10 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
2140 |
0.3 |
| chr4_84034679_84034947 | 0.10 |
PLAC8 |
placenta-specific 8 |
1055 |
0.6 |
| chr4_26270650_26270801 | 0.10 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
3504 |
0.38 |
| chr17_40405293_40405444 | 0.10 |
RP11-358B23.5 |
|
15237 |
0.11 |
| chrX_15756367_15757484 | 0.10 |
CA5B |
carbonic anhydrase VB, mitochondrial |
532 |
0.77 |
| chr6_42739563_42739714 | 0.10 |
GLTSCR1L |
GLTSCR1-like |
10127 |
0.17 |
| chr2_70321192_70321429 | 0.10 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
5332 |
0.17 |
| chr13_52108709_52108900 | 0.10 |
INTS6-AS1 |
INTS6 antisense RNA 1 |
6890 |
0.15 |
| chrX_130897041_130897446 | 0.10 |
ENSG00000200587 |
. |
175741 |
0.03 |
| chr1_32713939_32714090 | 0.10 |
FAM167B |
family with sequence similarity 167, member B |
1180 |
0.26 |
| chr11_2321078_2321236 | 0.10 |
C11orf21 |
chromosome 11 open reading frame 21 |
1986 |
0.2 |
| chr1_25890351_25890502 | 0.10 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
20355 |
0.18 |
| chr11_67172347_67172531 | 0.10 |
TBC1D10C |
TBC1 domain family, member 10C |
779 |
0.35 |
| chr17_6560768_6561014 | 0.10 |
ENSG00000264468 |
. |
2063 |
0.17 |
| chr14_22538566_22539116 | 0.10 |
ENSG00000238634 |
. |
72046 |
0.12 |
| chr8_29324075_29324226 | 0.10 |
RP4-676L2.1 |
|
113463 |
0.06 |
| chr8_82022969_82023806 | 0.10 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
916 |
0.72 |
| chr12_69201397_69201692 | 0.10 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
412 |
0.8 |
| chr17_62982296_62982856 | 0.09 |
AMZ2P1 |
archaelysin family metallopeptidase 2 pseudogene 1 |
12941 |
0.15 |
| chr13_114875917_114876068 | 0.09 |
RASA3-IT1 |
RASA3 intronic transcript 1 (non-protein coding) |
1697 |
0.42 |
| chr15_77289158_77289399 | 0.09 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
1363 |
0.44 |
| chr7_7812087_7812527 | 0.09 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
315 |
0.93 |
| chr6_143228096_143228637 | 0.09 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
37972 |
0.2 |
| chr18_48680545_48680786 | 0.09 |
MEX3C |
mex-3 RNA binding family member C |
42464 |
0.16 |
| chr1_198648140_198648351 | 0.09 |
RP11-553K8.5 |
|
12055 |
0.25 |
| chr5_39197523_39197702 | 0.09 |
FYB |
FYN binding protein |
5517 |
0.3 |
| chr8_28559464_28559708 | 0.09 |
EXTL3-AS1 |
EXTL3 antisense RNA 1 |
605 |
0.51 |
| chr7_8010738_8010889 | 0.09 |
AC006042.7 |
|
1279 |
0.36 |
| chrY_1605029_1605190 | 0.09 |
NA |
NA |
> 106 |
NA |
| chr1_28195799_28196249 | 0.09 |
THEMIS2 |
thymocyte selection associated family member 2 |
3031 |
0.15 |
| chr13_103498299_103498896 | 0.09 |
ERCC5 |
excision repair cross-complementing rodent repair deficiency, complementation group 5 |
423 |
0.79 |
| chr1_6663176_6663786 | 0.09 |
KLHL21 |
kelch-like family member 21 |
420 |
0.75 |
| chr1_204379460_204380399 | 0.09 |
PPP1R15B |
protein phosphatase 1, regulatory subunit 15B |
990 |
0.54 |
| chr1_29242251_29242585 | 0.09 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
1327 |
0.43 |
| chrX_117632459_117632610 | 0.09 |
DOCK11 |
dedicator of cytokinesis 11 |
2662 |
0.35 |
| chr1_144340215_144340417 | 0.09 |
AL592284.1 |
Protein LOC642441 |
578 |
0.76 |
| chrX_153166301_153166452 | 0.09 |
AVPR2 |
arginine vasopressin receptor 2 |
3813 |
0.11 |
| chr4_144480155_144480513 | 0.09 |
SMARCA5-AS1 |
SMARCA5 antisense RNA 1 |
44546 |
0.15 |
| chrX_1655035_1655190 | 0.09 |
P2RY8 |
purinergic receptor P2Y, G-protein coupled, 8 |
888 |
0.64 |
| chr9_134143747_134144006 | 0.09 |
FAM78A |
family with sequence similarity 78, member A |
2004 |
0.31 |
| chr3_114009980_114010222 | 0.09 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
152 |
0.96 |
| chr12_92899150_92899489 | 0.09 |
ENSG00000238865 |
. |
39444 |
0.17 |
| chr1_47903753_47903904 | 0.09 |
FOXD2 |
forkhead box D2 |
2139 |
0.25 |
| chr8_58504015_58504166 | 0.09 |
ENSG00000252057 |
. |
34946 |
0.24 |
| chr6_143228681_143228886 | 0.09 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
37555 |
0.2 |
| chr17_2700217_2700494 | 0.09 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
579 |
0.74 |
| chr5_75627068_75627317 | 0.09 |
RP11-466P24.6 |
|
19905 |
0.25 |
| chr20_62368079_62368369 | 0.09 |
LIME1 |
Lck interacting transmembrane adaptor 1 |
229 |
0.81 |
| chr19_16438433_16439148 | 0.09 |
KLF2 |
Kruppel-like factor 2 |
3139 |
0.19 |
| chr11_107879361_107879512 | 0.09 |
CUL5 |
cullin 5 |
23 |
0.98 |
| chr19_5804658_5804817 | 0.09 |
DUS3L |
dihydrouridine synthase 3-like (S. cerevisiae) |
13488 |
0.08 |
| chr1_151042732_151043215 | 0.09 |
GABPB2 |
GA binding protein transcription factor, beta subunit 2 |
107 |
0.58 |
| chr20_43599792_43600332 | 0.09 |
STK4 |
serine/threonine kinase 4 |
4895 |
0.16 |
| chr19_4379487_4379638 | 0.09 |
SH3GL1 |
SH3-domain GRB2-like 1 |
727 |
0.43 |
| chr22_39547258_39547409 | 0.09 |
CBX7 |
chromobox homolog 7 |
1116 |
0.46 |
| chr14_22962722_22962873 | 0.09 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
6626 |
0.11 |
| chr20_42840227_42840385 | 0.09 |
OSER1 |
oxidative stress responsive serine-rich 1 |
875 |
0.57 |
| chr1_182289672_182289823 | 0.09 |
ENSG00000206764 |
. |
6456 |
0.27 |
| chr13_19174246_19174726 | 0.09 |
ENSG00000223024 |
. |
268945 |
0.02 |
| chr19_7723134_7723321 | 0.09 |
RETN |
resistin |
10703 |
0.07 |
| chr15_63502851_63503002 | 0.09 |
RAB8B |
RAB8B, member RAS oncogene family |
21121 |
0.19 |
| chr10_63753311_63753523 | 0.09 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
55553 |
0.14 |
| chr19_10397706_10398023 | 0.09 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
184 |
0.83 |
| chr19_55765720_55766271 | 0.09 |
PPP6R1 |
protein phosphatase 6, regulatory subunit 1 |
1142 |
0.27 |
| chr6_26366833_26367042 | 0.09 |
ENSG00000215979 |
. |
1157 |
0.26 |
| chr19_13215586_13215874 | 0.09 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
1755 |
0.21 |
| chr6_117870110_117870478 | 0.09 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
53233 |
0.12 |
| chr5_176839896_176840051 | 0.09 |
F12 |
coagulation factor XII (Hageman factor) |
3396 |
0.12 |
| chr1_249156955_249157464 | 0.09 |
AL672294.1 |
|
3846 |
0.16 |
| chr17_39822401_39822869 | 0.09 |
EIF1 |
eukaryotic translation initiation factor 1 |
22504 |
0.09 |
| chr19_54873932_54874135 | 0.09 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
1477 |
0.26 |
| chr19_17982701_17982908 | 0.09 |
SLC5A5 |
solute carrier family 5 (sodium/iodide cotransporter), member 5 |
22 |
0.96 |
| chr14_52327497_52328482 | 0.09 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
53 |
0.98 |
| chr2_43448437_43448588 | 0.09 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
5236 |
0.25 |
| chr5_118644851_118645262 | 0.09 |
ENSG00000243333 |
. |
2730 |
0.28 |
| chr20_3779530_3779768 | 0.09 |
CDC25B |
cell division cycle 25B |
2571 |
0.18 |
| chr9_126101731_126102049 | 0.09 |
CRB2 |
crumbs homolog 2 (Drosophila) |
16559 |
0.22 |
| chr2_242557475_242557626 | 0.09 |
THAP4 |
THAP domain containing 4 |
634 |
0.62 |
| chr19_9937977_9938220 | 0.09 |
FBXL12 |
F-box and leucine-rich repeat protein 12 |
394 |
0.46 |
| chr16_14380439_14380590 | 0.09 |
ENSG00000201075 |
. |
11152 |
0.18 |
| chr3_182511391_182511643 | 0.09 |
ATP11B |
ATPase, class VI, type 11B |
226 |
0.63 |
| chr4_3304601_3305007 | 0.09 |
RGS12 |
regulator of G-protein signaling 12 |
5958 |
0.26 |
| chr6_128293411_128293648 | 0.08 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
35693 |
0.18 |
| chr4_122103316_122103738 | 0.08 |
ENSG00000252183 |
. |
10531 |
0.22 |
| chr9_32572870_32573104 | 0.08 |
NDUFB6 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa |
170 |
0.93 |
| chr16_30220301_30220755 | 0.08 |
RP11-347C12.3 |
Uncharacterized protein |
2025 |
0.15 |
| chr1_6525547_6525781 | 0.08 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
26 |
0.96 |
| chr12_50064807_50065013 | 0.08 |
FMNL3 |
formin-like 3 |
36093 |
0.11 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.3 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.0 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.2 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.0 | 0.2 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:1901538 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.2 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.1 | GO:1903313 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.5 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0002677 | negative regulation of chronic inflammatory response(GO:0002677) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.3 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0033233 | regulation of protein sumoylation(GO:0033233) positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.7 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.0 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.1 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.0 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.0 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0032608 | interferon-beta production(GO:0032608) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0032727 | positive regulation of interferon-alpha production(GO:0032727) |
| 0.0 | 0.1 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0021612 | cranial nerve structural organization(GO:0021604) facial nerve morphogenesis(GO:0021610) facial nerve structural organization(GO:0021612) anatomical structure arrangement(GO:0048532) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0019908 | nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.9 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.0 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.0 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.1 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.0 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.1 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |