Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
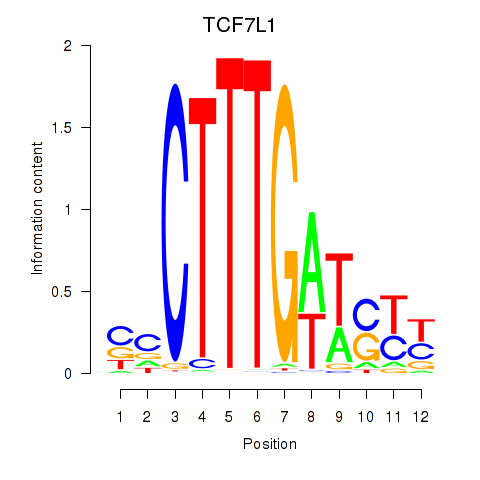
Results for TCF7L1
Z-value: 0.70
Transcription factors associated with TCF7L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TCF7L1
|
ENSG00000152284.4 | transcription factor 7 like 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_85443412_85443563 | TCF7L1 | 2741 | 0.220079 | -0.83 | 6.1e-03 | Click! |
| chr2_85465249_85465400 | TCF7L1 | 24578 | 0.125901 | 0.75 | 2.1e-02 | Click! |
| chr2_85464644_85464795 | TCF7L1 | 23973 | 0.127088 | 0.70 | 3.5e-02 | Click! |
| chr2_85476158_85476514 | TCF7L1 | 35590 | 0.103205 | -0.54 | 1.3e-01 | Click! |
| chr2_85473701_85473852 | TCF7L1 | 33030 | 0.108565 | 0.53 | 1.4e-01 | Click! |
Activity of the TCF7L1 motif across conditions
Conditions sorted by the z-value of the TCF7L1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
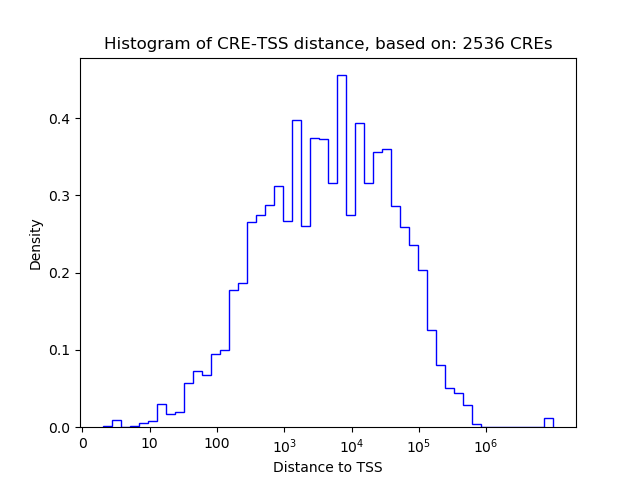
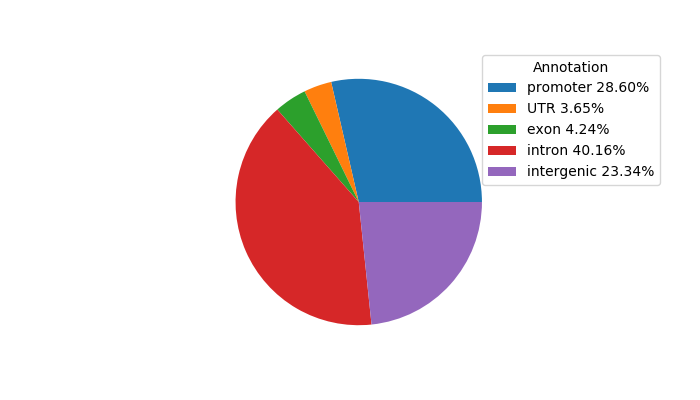
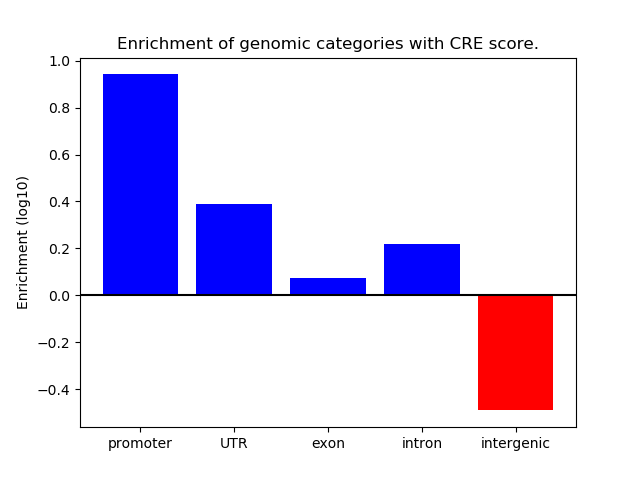
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_80279192_80279518 | 0.28 |
CD7 |
CD7 molecule |
3877 |
0.13 |
| chr7_27184642_27184864 | 0.26 |
HOXA5 |
homeobox A5 |
1466 |
0.14 |
| chr8_17611273_17611424 | 0.25 |
MTUS1 |
microtubule associated tumor suppressor 1 |
2142 |
0.28 |
| chr16_9192642_9192817 | 0.25 |
RP11-473I1.9 |
|
5976 |
0.16 |
| chr4_153194568_153194863 | 0.25 |
ENSG00000244544 |
. |
9118 |
0.23 |
| chr1_192514118_192514603 | 0.23 |
RP5-1011O1.2 |
|
21987 |
0.19 |
| chr1_95015118_95015269 | 0.23 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
7837 |
0.3 |
| chr7_100223660_100223811 | 0.22 |
TFR2 |
transferrin receptor 2 |
6992 |
0.09 |
| chr10_132972880_132973031 | 0.21 |
TCERG1L-AS1 |
TCERG1L antisense RNA 1 |
79474 |
0.11 |
| chr10_113590379_113590912 | 0.20 |
GPAM |
glycerol-3-phosphate acyltransferase, mitochondrial |
352823 |
0.01 |
| chr15_66083482_66084093 | 0.20 |
DENND4A |
DENN/MADD domain containing 4A |
675 |
0.68 |
| chr16_2975177_2975328 | 0.20 |
FLYWCH1 |
FLYWCH-type zinc finger 1 |
5546 |
0.1 |
| chr1_33428156_33429049 | 0.19 |
RNF19B |
ring finger protein 19B |
1684 |
0.28 |
| chr5_175918974_175919125 | 0.19 |
RNF44 |
ring finger protein 44 |
38597 |
0.09 |
| chr9_79637286_79637437 | 0.19 |
FOXB2 |
forkhead box B2 |
2790 |
0.39 |
| chr1_159929593_159929744 | 0.19 |
SLAMF9 |
SLAM family member 9 |
5624 |
0.12 |
| chr3_73126790_73126941 | 0.19 |
ENSG00000238416 |
. |
4273 |
0.16 |
| chr3_149375959_149376254 | 0.19 |
WWTR1-AS1 |
WWTR1 antisense RNA 1 |
109 |
0.66 |
| chr6_130543841_130543992 | 0.18 |
SAMD3 |
sterile alpha motif domain containing 3 |
42 |
0.99 |
| chr19_30842754_30842905 | 0.18 |
ZNF536 |
zinc finger protein 536 |
20490 |
0.28 |
| chr1_41966637_41966788 | 0.18 |
EDN2 |
endothelin 2 |
16370 |
0.19 |
| chr17_63694359_63694510 | 0.18 |
CEP112 |
centrosomal protein 112kDa |
52408 |
0.18 |
| chrX_13185584_13185735 | 0.18 |
FAM9C |
family with sequence similarity 9, member C |
122858 |
0.05 |
| chr2_100102004_100102155 | 0.18 |
REV1 |
REV1, polymerase (DNA directed) |
4356 |
0.25 |
| chr19_34669041_34669419 | 0.18 |
LSM14A |
LSM14A, SCD6 homolog A (S. cerevisiae) |
5667 |
0.28 |
| chr20_32613579_32613730 | 0.17 |
RALY |
RALY heterogeneous nuclear ribonucleoprotein |
4013 |
0.23 |
| chr14_98443744_98443895 | 0.17 |
C14orf64 |
chromosome 14 open reading frame 64 |
564 |
0.87 |
| chr8_144576890_144577041 | 0.17 |
ZC3H3 |
zinc finger CCCH-type containing 3 |
13045 |
0.1 |
| chr7_150180608_150181504 | 0.17 |
GIMAP7 |
GTPase, IMAP family member 7 |
30862 |
0.13 |
| chr14_106000080_106000231 | 0.17 |
TMEM121 |
transmembrane protein 121 |
5194 |
0.14 |
| chr6_144672449_144672906 | 0.17 |
UTRN |
utrophin |
7440 |
0.26 |
| chr20_15982068_15982219 | 0.17 |
MACROD2 |
MACRO domain containing 2 |
15710 |
0.31 |
| chr7_139929395_139929688 | 0.16 |
ENSG00000199283 |
. |
12129 |
0.17 |
| chr1_160680712_160681035 | 0.16 |
CD48 |
CD48 molecule |
720 |
0.63 |
| chr12_10328213_10328364 | 0.16 |
TMEM52B |
transmembrane protein 52B |
3324 |
0.16 |
| chr11_108724912_108725630 | 0.16 |
ENSG00000201243 |
. |
150973 |
0.04 |
| chr1_198621955_198622106 | 0.16 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
13738 |
0.23 |
| chr2_192169808_192170004 | 0.16 |
MYO1B |
myosin IB |
28295 |
0.22 |
| chr10_22625053_22625204 | 0.16 |
SPAG6 |
sperm associated antigen 6 |
9271 |
0.15 |
| chr3_13919959_13920372 | 0.16 |
WNT7A |
wingless-type MMTV integration site family, member 7A |
1453 |
0.48 |
| chr9_135506565_135506716 | 0.16 |
DDX31 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
32247 |
0.15 |
| chr3_129468296_129468447 | 0.15 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
44888 |
0.13 |
| chr11_130258989_130259140 | 0.15 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
39824 |
0.16 |
| chr5_132377333_132377484 | 0.15 |
HSPA4 |
heat shock 70kDa protein 4 |
10246 |
0.17 |
| chr2_236768694_236768845 | 0.15 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
23270 |
0.24 |
| chrX_77204120_77204271 | 0.15 |
PGAM4 |
phosphoglycerate mutase family member 4 |
20940 |
0.16 |
| chr3_66514964_66515132 | 0.15 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
36308 |
0.22 |
| chr1_2384159_2384310 | 0.15 |
PLCH2 |
phospholipase C, eta 2 |
14664 |
0.11 |
| chr2_210241109_210241260 | 0.15 |
MAP2 |
microtubule-associated protein 2 |
47598 |
0.2 |
| chr6_135504304_135504969 | 0.15 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
1984 |
0.32 |
| chr17_38264116_38264342 | 0.15 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
7251 |
0.12 |
| chr11_134145279_134145568 | 0.15 |
GLB1L3 |
galactosidase, beta 1-like 3 |
1212 |
0.41 |
| chr20_62573788_62573939 | 0.14 |
ENSG00000207554 |
. |
216 |
0.73 |
| chr10_61624489_61624640 | 0.14 |
CCDC6 |
coiled-coil domain containing 6 |
41850 |
0.19 |
| chr7_129419391_129420014 | 0.14 |
ENSG00000207691 |
. |
4848 |
0.17 |
| chr14_22987127_22987278 | 0.14 |
TRAJ15 |
T cell receptor alpha joining 15 |
11378 |
0.1 |
| chr12_6744815_6744999 | 0.14 |
LPAR5 |
lysophosphatidic acid receptor 5 |
706 |
0.45 |
| chr10_13732369_13732520 | 0.14 |
RP11-295P9.8 |
|
7923 |
0.13 |
| chr20_56193089_56193240 | 0.14 |
ZBP1 |
Z-DNA binding protein 1 |
2286 |
0.35 |
| chr4_7971561_7971846 | 0.14 |
AFAP1 |
actin filament associated protein 1 |
30050 |
0.14 |
| chr6_12061250_12061691 | 0.14 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
45650 |
0.18 |
| chrY_14081502_14081653 | 0.14 |
ENSG00000265161 |
. |
134065 |
0.06 |
| chr11_111749891_111750174 | 0.14 |
C11orf1 |
chromosome 11 open reading frame 1 |
84 |
0.55 |
| chr2_58250808_58250969 | 0.14 |
VRK2 |
vaccinia related kinase 2 |
22841 |
0.23 |
| chr17_37607615_37607816 | 0.14 |
MED1 |
mediator complex subunit 1 |
176 |
0.93 |
| chr22_37310111_37310410 | 0.14 |
CSF2RB |
colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
585 |
0.66 |
| chr10_4728192_4728343 | 0.14 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
100554 |
0.08 |
| chr1_89487830_89488505 | 0.14 |
GBP3 |
guanylate binding protein 3 |
382 |
0.8 |
| chr12_40012002_40012194 | 0.14 |
ABCD2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
1455 |
0.48 |
| chr1_154979820_154980060 | 0.14 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
4645 |
0.08 |
| chr15_70250319_70250470 | 0.14 |
ENSG00000207965 |
. |
121413 |
0.06 |
| chr10_124269483_124269634 | 0.13 |
HTRA1 |
HtrA serine peptidase 1 |
3351 |
0.27 |
| chr17_28553116_28553267 | 0.13 |
SLC6A4 |
solute carrier family 6 (neurotransmitter transporter), member 4 |
9525 |
0.11 |
| chr5_1884051_1884202 | 0.13 |
CTD-2194D22.3 |
|
46 |
0.95 |
| chr14_69158161_69158312 | 0.13 |
CTD-2325P2.3 |
|
5954 |
0.24 |
| chr7_103273793_103274056 | 0.13 |
ENSG00000222386 |
. |
148938 |
0.04 |
| chr6_29932866_29933025 | 0.13 |
HLA-A |
major histocompatibility complex, class I, A |
22619 |
0.14 |
| chr9_134153488_134153764 | 0.13 |
FAM78A |
family with sequence similarity 78, member A |
1692 |
0.35 |
| chrX_154417087_154417278 | 0.13 |
VBP1 |
von Hippel-Lindau binding protein 1 |
8102 |
0.19 |
| chr2_121716938_121717089 | 0.13 |
ENSG00000221321 |
. |
75636 |
0.11 |
| chr11_44591831_44591982 | 0.13 |
CD82 |
CD82 molecule |
2722 |
0.31 |
| chr10_30675999_30676150 | 0.13 |
ENSG00000239625 |
. |
20827 |
0.15 |
| chr11_64641916_64642067 | 0.13 |
EHD1 |
EH-domain containing 1 |
1147 |
0.31 |
| chr1_155101296_155101447 | 0.13 |
EFNA1 |
ephrin-A1 |
1019 |
0.27 |
| chr13_46753488_46753737 | 0.13 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
2847 |
0.22 |
| chr19_59071940_59072091 | 0.13 |
AC016629.8 |
|
1423 |
0.18 |
| chrX_40013569_40013864 | 0.12 |
BCOR |
BCL6 corepressor |
7834 |
0.32 |
| chr19_461771_462005 | 0.12 |
SHC2 |
SHC (Src homology 2 domain containing) transforming protein 2 |
892 |
0.42 |
| chr6_55501172_55501323 | 0.12 |
HMGCLL1 |
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
57235 |
0.17 |
| chr16_30818504_30818674 | 0.12 |
ZNF629 |
zinc finger protein 629 |
20066 |
0.08 |
| chr8_70745749_70746362 | 0.12 |
RP11-159H10.3 |
|
291 |
0.62 |
| chr10_30017356_30017507 | 0.12 |
SVIL |
supervillin |
7299 |
0.24 |
| chr9_101011054_101011278 | 0.12 |
TBC1D2 |
TBC1 domain family, member 2 |
6696 |
0.27 |
| chr19_43100043_43100194 | 0.12 |
CEACAM8 |
carcinoembryonic antigen-related cell adhesion molecule 8 |
911 |
0.61 |
| chr16_89382358_89382591 | 0.12 |
AC137932.6 |
|
5067 |
0.15 |
| chr17_15851287_15851438 | 0.12 |
ADORA2B |
adenosine A2b receptor |
3131 |
0.25 |
| chr13_76334632_76334929 | 0.12 |
LMO7 |
LIM domain 7 |
15 |
0.99 |
| chr21_43648519_43648670 | 0.12 |
ENSG00000223262 |
. |
7311 |
0.18 |
| chr10_22622036_22622322 | 0.12 |
BMI1 |
BMI1 polycomb ring finger oncogene |
6731 |
0.15 |
| chr7_100488371_100488522 | 0.12 |
UFSP1 |
UFM1-specific peptidase 1 (non-functional) |
1107 |
0.3 |
| chr15_52338021_52338172 | 0.11 |
RP11-430B1.1 |
|
12192 |
0.15 |
| chr12_43681939_43682090 | 0.11 |
ENSG00000215993 |
. |
3369 |
0.39 |
| chr21_39672293_39672488 | 0.11 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
3542 |
0.35 |
| chr3_71775115_71775529 | 0.11 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
796 |
0.68 |
| chr20_31100174_31100325 | 0.11 |
C20orf112 |
chromosome 20 open reading frame 112 |
1039 |
0.53 |
| chr1_111212990_111213454 | 0.11 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
4433 |
0.21 |
| chr11_95187135_95187286 | 0.11 |
ENSG00000201204 |
. |
19328 |
0.28 |
| chr11_128381553_128381704 | 0.11 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
6339 |
0.25 |
| chr1_24828022_24828241 | 0.11 |
RCAN3 |
RCAN family member 3 |
1256 |
0.44 |
| chr19_16195500_16195773 | 0.11 |
TPM4 |
tropomyosin 4 |
3103 |
0.21 |
| chr7_41922063_41922214 | 0.11 |
AC005027.3 |
|
177205 |
0.03 |
| chr19_4911213_4911907 | 0.11 |
ARRDC5 |
arrestin domain containing 5 |
8681 |
0.13 |
| chr7_150655998_150656149 | 0.11 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
1891 |
0.27 |
| chr4_177366271_177366422 | 0.11 |
RP11-87F15.2 |
|
124737 |
0.05 |
| chr1_210407355_210407517 | 0.11 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
44 |
0.97 |
| chr2_204734419_204734570 | 0.11 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
461 |
0.87 |
| chr1_109757024_109757692 | 0.11 |
SARS |
seryl-tRNA synthetase |
754 |
0.58 |
| chr5_32531847_32531998 | 0.11 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
2 |
0.99 |
| chr3_71834842_71834993 | 0.11 |
PROK2 |
prokineticin 2 |
560 |
0.77 |
| chr11_64088050_64088201 | 0.11 |
PRDX5 |
peroxiredoxin 5 |
2471 |
0.1 |
| chr14_50988685_50989139 | 0.11 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
10278 |
0.18 |
| chr13_26042483_26042809 | 0.11 |
ATP8A2 |
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
467 |
0.84 |
| chr1_158624437_158624588 | 0.11 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
31886 |
0.12 |
| chr5_171580625_171580776 | 0.11 |
STK10 |
serine/threonine kinase 10 |
34690 |
0.15 |
| chr3_171774780_171774931 | 0.11 |
FNDC3B |
fibronectin type III domain containing 3B |
10152 |
0.29 |
| chr5_112497806_112497957 | 0.11 |
MCC |
mutated in colorectal cancers |
72493 |
0.11 |
| chr1_154955826_154956219 | 0.11 |
FLAD1 |
flavin adenine dinucleotide synthetase 1 |
173 |
0.86 |
| chr7_45151465_45151735 | 0.11 |
TBRG4 |
transforming growth factor beta regulator 4 |
20 |
0.96 |
| chr1_24882080_24882472 | 0.11 |
NCMAP |
noncompact myelin associated protein |
326 |
0.88 |
| chr15_59576524_59576675 | 0.11 |
RP11-429D19.1 |
|
13238 |
0.15 |
| chr17_39778090_39778241 | 0.10 |
KRT17 |
keratin 17 |
1355 |
0.27 |
| chr7_76149676_76149827 | 0.10 |
ENSG00000238560 |
. |
3171 |
0.19 |
| chr22_46971721_46971872 | 0.10 |
GRAMD4 |
GRAM domain containing 4 |
113 |
0.97 |
| chr1_52363576_52363890 | 0.10 |
ENSG00000265769 |
. |
2910 |
0.2 |
| chr17_75447598_75448179 | 0.10 |
SEPT9 |
septin 9 |
562 |
0.71 |
| chr7_93485635_93485786 | 0.10 |
TFPI2 |
tissue factor pathway inhibitor 2 |
33772 |
0.15 |
| chr10_3542796_3543180 | 0.10 |
RP11-184A2.3 |
|
250271 |
0.02 |
| chr7_138619859_138620309 | 0.10 |
KIAA1549 |
KIAA1549 |
15863 |
0.22 |
| chr22_45714336_45714978 | 0.10 |
FAM118A |
family with sequence similarity 118, member A |
15 |
0.98 |
| chr1_2981320_2981471 | 0.10 |
LINC00982 |
long intergenic non-protein coding RNA 982 |
1045 |
0.53 |
| chr2_60758991_60759142 | 0.10 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
21474 |
0.21 |
| chr3_174486803_174486954 | 0.10 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
90192 |
0.1 |
| chr2_36796670_36797008 | 0.10 |
FEZ2 |
fasciculation and elongation protein zeta 2 (zygin II) |
11631 |
0.19 |
| chr12_27717509_27717879 | 0.10 |
PPFIBP1 |
PTPRF interacting protein, binding protein 1 (liprin beta 1) |
26638 |
0.2 |
| chr17_7297534_7297804 | 0.10 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
76 |
0.9 |
| chr15_55559668_55560071 | 0.10 |
RAB27A |
RAB27A, member RAS oncogene family |
2585 |
0.3 |
| chr2_139273768_139273919 | 0.10 |
SPOPL |
speckle-type POZ protein-like |
14472 |
0.31 |
| chr7_55637667_55637921 | 0.10 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
1767 |
0.37 |
| chr10_29887628_29887779 | 0.10 |
ENSG00000216035 |
. |
3572 |
0.24 |
| chr8_8154312_8154463 | 0.10 |
ALG1L13P |
asparagine-linked glycosylation 1-like 13, pseudogene |
55454 |
0.11 |
| chr2_106711904_106712070 | 0.10 |
C2orf40 |
chromosome 2 open reading frame 40 |
29731 |
0.2 |
| chr5_176782074_176782225 | 0.10 |
RGS14 |
regulator of G-protein signaling 14 |
2689 |
0.14 |
| chr9_12241558_12241709 | 0.10 |
ENSG00000222581 |
. |
58818 |
0.17 |
| chr17_57643262_57643540 | 0.10 |
DHX40 |
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
417 |
0.85 |
| chr2_192176773_192176924 | 0.10 |
MYO1B |
myosin IB |
35237 |
0.19 |
| chr18_3453699_3454909 | 0.10 |
TGIF1 |
TGFB-induced factor homeobox 1 |
532 |
0.8 |
| chrX_19815653_19816006 | 0.10 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
2040 |
0.46 |
| chr2_204801338_204802214 | 0.10 |
ICOS |
inducible T-cell co-stimulator |
273 |
0.95 |
| chr19_16230354_16230505 | 0.10 |
RAB8A |
RAB8A, member RAS oncogene family |
7722 |
0.13 |
| chr9_23779204_23779355 | 0.10 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
94 |
0.99 |
| chr16_3628507_3629057 | 0.10 |
NLRC3 |
NLR family, CARD domain containing 3 |
1381 |
0.35 |
| chr8_144591297_144591448 | 0.10 |
ZC3H3 |
zinc finger CCCH-type containing 3 |
1362 |
0.27 |
| chr2_45970517_45970668 | 0.10 |
U51244.2 |
|
67977 |
0.12 |
| chr1_205287839_205287990 | 0.10 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
2969 |
0.23 |
| chr17_80832203_80832354 | 0.10 |
TBCD |
tubulin folding cofactor D |
9750 |
0.18 |
| chr10_54213744_54213895 | 0.10 |
RP11-556E13.1 |
|
107046 |
0.07 |
| chr17_53785965_53786128 | 0.10 |
TMEM100 |
transmembrane protein 100 |
13113 |
0.26 |
| chrY_9769735_9769886 | 0.09 |
TSPY5P |
testis specific protein, Y-linked 5, pseudogene |
136950 |
0.05 |
| chr11_117857636_117858098 | 0.09 |
IL10RA |
interleukin 10 receptor, alpha |
758 |
0.66 |
| chr18_11688870_11689119 | 0.09 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
39 |
0.98 |
| chr17_54671562_54672245 | 0.09 |
NOG |
noggin |
843 |
0.75 |
| chrY_21729956_21730145 | 0.09 |
TXLNG2P |
taxilin gamma 2, pseudogene |
782 |
0.7 |
| chr8_22524109_22524260 | 0.09 |
BIN3 |
bridging integrator 3 |
2442 |
0.18 |
| chr16_84875483_84875757 | 0.09 |
CRISPLD2 |
cysteine-rich secretory protein LCCL domain containing 2 |
11 |
0.98 |
| chr8_17105056_17105444 | 0.09 |
VPS37A |
vacuolar protein sorting 37 homolog A (S. cerevisiae) |
497 |
0.64 |
| chr5_167249979_167250130 | 0.09 |
ENSG00000221741 |
. |
47616 |
0.14 |
| chr2_122257749_122257900 | 0.09 |
CLASP1 |
cytoplasmic linker associated protein 1 |
3740 |
0.25 |
| chr5_133457391_133457692 | 0.09 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
1768 |
0.4 |
| chr7_150414678_150414897 | 0.09 |
GIMAP1 |
GTPase, IMAP family member 1 |
1142 |
0.45 |
| chr21_47285255_47285499 | 0.09 |
PCBP3 |
poly(rC) binding protein 3 |
15502 |
0.19 |
| chr17_61328641_61328792 | 0.09 |
AC037445.1 |
|
53570 |
0.11 |
| chr4_41938007_41938269 | 0.09 |
TMEM33 |
transmembrane protein 33 |
969 |
0.62 |
| chr4_53058800_53058951 | 0.09 |
SPATA18 |
spermatogenesis associated 18 |
141293 |
0.05 |
| chr4_40196291_40196444 | 0.09 |
RHOH |
ras homolog family member H |
1084 |
0.55 |
| chr1_86874016_86874167 | 0.09 |
ODF2L |
outer dense fiber of sperm tails 2-like |
12068 |
0.21 |
| chr2_119067816_119068522 | 0.09 |
INSIG2 |
insulin induced gene 2 |
222119 |
0.02 |
| chr6_130197055_130197206 | 0.09 |
TMEM244 |
transmembrane protein 244 |
14438 |
0.28 |
| chr17_56401068_56401219 | 0.09 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
1668 |
0.23 |
| chr1_155053219_155053398 | 0.09 |
EFNA3 |
ephrin-A3 |
1923 |
0.15 |
| chr17_38717958_38718109 | 0.09 |
CCR7 |
chemokine (C-C motif) receptor 7 |
768 |
0.63 |
| chr3_111851417_111851568 | 0.09 |
GCSAM |
germinal center-associated, signaling and motility |
580 |
0.52 |
| chrX_153359472_153359623 | 0.09 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
2889 |
0.2 |
| chr7_27198059_27198210 | 0.09 |
HOXA7 |
homeobox A7 |
579 |
0.44 |
| chr19_54384929_54385717 | 0.09 |
PRKCG |
protein kinase C, gamma |
130 |
0.89 |
| chr17_74495503_74495654 | 0.09 |
RHBDF2 |
rhomboid 5 homolog 2 (Drosophila) |
1875 |
0.2 |
| chr8_29513452_29513654 | 0.09 |
ENSG00000221003 |
. |
272568 |
0.01 |
| chr11_5246641_5246814 | 0.09 |
ENSG00000221031 |
. |
858 |
0.32 |
| chr16_66551368_66551754 | 0.09 |
TK2 |
thymidine kinase 2, mitochondrial |
3875 |
0.13 |
| chr20_30104549_30104700 | 0.09 |
HM13 |
histocompatibility (minor) 13 |
2300 |
0.21 |
| chr17_66508156_66508438 | 0.09 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
124 |
0.97 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.1 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.0 | 0.1 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.0 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.0 | GO:0060087 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.0 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |