Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
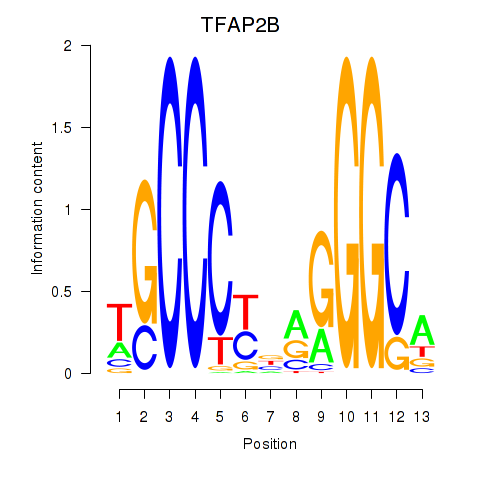
Results for TFAP2B
Z-value: 1.36
Transcription factors associated with TFAP2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TFAP2B
|
ENSG00000008196.8 | transcription factor AP-2 beta |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_50812700_50812851 | TFAP2B | 26336 | 0.267431 | 0.79 | 1.1e-02 | Click! |
| chr6_50788009_50788160 | TFAP2B | 1645 | 0.543666 | 0.73 | 2.5e-02 | Click! |
| chr6_50811539_50811690 | TFAP2B | 25175 | 0.271090 | 0.67 | 5.0e-02 | Click! |
| chr6_50817900_50818051 | TFAP2B | 31536 | 0.250523 | 0.65 | 5.9e-02 | Click! |
| chr6_50810973_50811124 | TFAP2B | 24609 | 0.272853 | 0.65 | 6.0e-02 | Click! |
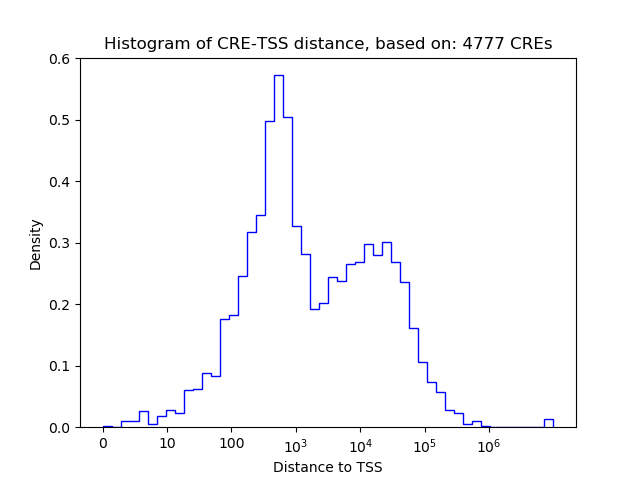
Activity of the TFAP2B motif across conditions
Conditions sorted by the z-value of the TFAP2B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
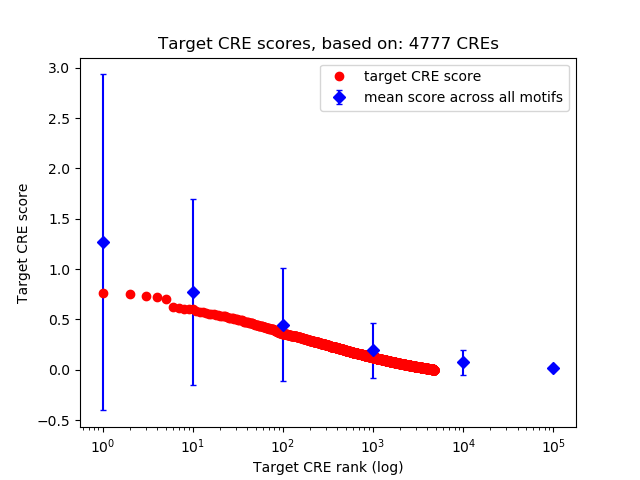
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_48712666_48713008 | 0.76 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
619 |
0.6 |
| chr1_34628677_34628828 | 0.75 |
CSMD2 |
CUB and Sushi multiple domains 2 |
2123 |
0.29 |
| chr1_956379_957022 | 0.73 |
AGRN |
agrin |
1197 |
0.27 |
| chr6_54711000_54712080 | 0.72 |
FAM83B |
family with sequence similarity 83, member B |
29 |
0.98 |
| chr22_44756680_44757093 | 0.70 |
RP1-32I10.10 |
Uncharacterized protein |
4545 |
0.29 |
| chr17_5371546_5372510 | 0.63 |
DHX33 |
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
40 |
0.96 |
| chr14_65171132_65172314 | 0.61 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
408 |
0.88 |
| chr8_22027174_22027581 | 0.61 |
BMP1 |
bone morphogenetic protein 1 |
4577 |
0.12 |
| chr7_150779695_150780926 | 0.60 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
9 |
0.94 |
| chr4_1035336_1035962 | 0.60 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
29410 |
0.11 |
| chr3_137486919_137487668 | 0.59 |
SOX14 |
SRY (sex determining region Y)-box 14 |
3714 |
0.37 |
| chr17_48137700_48137955 | 0.58 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
4023 |
0.16 |
| chr19_5339568_5339818 | 0.57 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
1121 |
0.59 |
| chr15_40531754_40531905 | 0.56 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
208 |
0.89 |
| chr5_131594297_131594770 | 0.56 |
PDLIM4 |
PDZ and LIM domain 4 |
1133 |
0.45 |
| chr12_112818867_112820166 | 0.56 |
HECTD4 |
HECT domain containing E3 ubiquitin protein ligase 4 |
380 |
0.87 |
| chr17_37895722_37895926 | 0.55 |
GRB7 |
growth factor receptor-bound protein 7 |
396 |
0.76 |
| chr2_241949003_241950045 | 0.54 |
AC093585.6 |
|
637 |
0.62 |
| chr15_99192776_99194432 | 0.54 |
IGF1R |
insulin-like growth factor 1 receptor |
1331 |
0.47 |
| chr3_53106938_53107738 | 0.54 |
ENSG00000266635 |
. |
20324 |
0.16 |
| chr3_128207705_128209147 | 0.54 |
RP11-475N22.4 |
|
326 |
0.79 |
| chr2_110872434_110873339 | 0.53 |
MALL |
mal, T-cell differentiation protein-like |
713 |
0.63 |
| chr4_187025490_187026623 | 0.53 |
FAM149A |
family with sequence similarity 149, member A |
175 |
0.94 |
| chr8_23260918_23261191 | 0.53 |
LOXL2 |
lysyl oxidase-like 2 |
535 |
0.78 |
| chr2_83156543_83156694 | 0.52 |
ENSG00000201311 |
. |
622326 |
0.0 |
| chr19_4584366_4584890 | 0.52 |
SEMA6B |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
24808 |
0.09 |
| chr4_164265662_164265856 | 0.52 |
NPY1R |
neuropeptide Y receptor Y1 |
225 |
0.76 |
| chr10_11784750_11785238 | 0.51 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
549 |
0.83 |
| chr19_51161641_51162497 | 0.50 |
C19orf81 |
chromosome 19 open reading frame 81 |
9024 |
0.12 |
| chr14_105330590_105330952 | 0.50 |
CEP170B |
centrosomal protein 170B |
846 |
0.6 |
| chr17_48423822_48424723 | 0.50 |
XYLT2 |
xylosyltransferase II |
776 |
0.5 |
| chr1_27286242_27286878 | 0.50 |
C1orf172 |
chromosome 1 open reading frame 172 |
337 |
0.84 |
| chr12_124907458_124907665 | 0.50 |
NCOR2 |
nuclear receptor corepressor 2 |
34191 |
0.22 |
| chr10_5637782_5638011 | 0.50 |
ENSG00000240577 |
. |
35530 |
0.13 |
| chr4_180979181_180979679 | 0.49 |
NA |
NA |
> 106 |
NA |
| chr6_5999099_5999932 | 0.48 |
NRN1 |
neuritin 1 |
7685 |
0.26 |
| chr6_169067989_169068140 | 0.48 |
SMOC2 |
SPARC related modular calcium binding 2 |
14300 |
0.31 |
| chr12_81471441_81472090 | 0.48 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
71 |
0.98 |
| chr2_87842220_87842459 | 0.48 |
RP11-1399P15.1 |
|
64786 |
0.13 |
| chr16_4986136_4987107 | 0.48 |
PPL |
periplakin |
444 |
0.78 |
| chr9_133302665_133302816 | 0.47 |
HMCN2 |
hemicentin 2 |
2929 |
0.21 |
| chr10_35102956_35103650 | 0.47 |
PARD3 |
par-3 family cell polarity regulator |
946 |
0.5 |
| chr15_74472398_74472549 | 0.47 |
RP11-665J16.1 |
|
906 |
0.44 |
| chr10_134659120_134659271 | 0.47 |
TTC40 |
tetratricopeptide repeat domain 40 |
10338 |
0.24 |
| chr1_204168084_204168306 | 0.46 |
KISS1 |
KiSS-1 metastasis-suppressor |
2581 |
0.22 |
| chr4_87769790_87770024 | 0.46 |
SLC10A6 |
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
509 |
0.85 |
| chr13_50985558_50985709 | 0.45 |
ENSG00000221198 |
. |
48260 |
0.19 |
| chr3_52016673_52017699 | 0.45 |
ABHD14B |
abhydrolase domain containing 14B |
239 |
0.49 |
| chr15_45455270_45455421 | 0.45 |
RP11-519G16.2 |
|
7197 |
0.1 |
| chr21_47394855_47395106 | 0.45 |
COL6A1 |
collagen, type VI, alpha 1 |
6671 |
0.21 |
| chr5_16934329_16934610 | 0.44 |
MYO10 |
myosin X |
1578 |
0.42 |
| chr3_138635529_138635680 | 0.44 |
FOXL2 |
forkhead box L2 |
30378 |
0.16 |
| chr1_242688162_242688638 | 0.44 |
PLD5 |
phospholipase D family, member 5 |
402 |
0.92 |
| chr2_46165643_46166035 | 0.44 |
PRKCE |
protein kinase C, epsilon |
62202 |
0.14 |
| chr5_172754207_172754714 | 0.44 |
STC2 |
stanniocalcin 2 |
596 |
0.75 |
| chr17_9018882_9019405 | 0.44 |
NTN1 |
netrin 1 |
47109 |
0.15 |
| chr1_151483990_151484782 | 0.44 |
CGN |
cingulin |
513 |
0.66 |
| chr5_3600266_3600509 | 0.43 |
CTD-2012M11.3 |
|
85 |
0.98 |
| chr1_201436884_201437966 | 0.43 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
887 |
0.55 |
| chr9_100849786_100850488 | 0.43 |
TRIM14 |
tripartite motif containing 14 |
4706 |
0.2 |
| chr22_19754354_19754802 | 0.42 |
TBX1 |
T-box 1 |
10352 |
0.18 |
| chr19_19216646_19216851 | 0.42 |
TMEM161A |
transmembrane protein 161A |
26462 |
0.11 |
| chr9_131013035_131013541 | 0.42 |
ENSG00000264823 |
. |
5979 |
0.09 |
| chr2_129365727_129365883 | 0.42 |
ENSG00000238379 |
. |
163033 |
0.04 |
| chr20_39970036_39970187 | 0.42 |
LPIN3 |
lipin 3 |
551 |
0.77 |
| chr16_1132922_1133073 | 0.42 |
SSTR5 |
somatostatin receptor 5 |
4128 |
0.16 |
| chr17_37856067_37857084 | 0.42 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
242 |
0.86 |
| chr5_17216444_17217195 | 0.41 |
AC091878.1 |
|
337 |
0.76 |
| chr2_27717607_27718081 | 0.41 |
FNDC4 |
fibronectin type III domain containing 4 |
268 |
0.82 |
| chr7_100464814_100466140 | 0.41 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr22_18506538_18507254 | 0.41 |
MICAL3 |
microtubule associated monooxygenase, calponin and LIM domain containing 3 |
429 |
0.79 |
| chr12_125002064_125002257 | 0.41 |
NCOR2 |
nuclear receptor corepressor 2 |
680 |
0.82 |
| chr2_133173748_133173945 | 0.40 |
GPR39 |
G protein-coupled receptor 39 |
301 |
0.92 |
| chr16_4422135_4423224 | 0.40 |
VASN |
vasorin |
830 |
0.49 |
| chr10_48354710_48355282 | 0.40 |
ZNF488 |
zinc finger protein 488 |
28 |
0.97 |
| chr19_41699238_41699877 | 0.40 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
405 |
0.76 |
| chr3_48631663_48631917 | 0.40 |
COL7A1 |
collagen, type VII, alpha 1 |
803 |
0.43 |
| chr9_130330127_130331202 | 0.40 |
FAM129B |
family with sequence similarity 129, member B |
703 |
0.67 |
| chr5_174871055_174871780 | 0.40 |
DRD1 |
dopamine receptor D1 |
206 |
0.95 |
| chr1_158969255_158969538 | 0.39 |
IFI16 |
interferon, gamma-inducible protein 16 |
362 |
0.89 |
| chr17_25879596_25879747 | 0.39 |
KSR1 |
kinase suppressor of ras 1 |
30276 |
0.17 |
| chr2_133427269_133427458 | 0.39 |
LYPD1 |
LY6/PLAUR domain containing 1 |
415 |
0.87 |
| chr15_40631412_40631563 | 0.39 |
C15orf52 |
chromosome 15 open reading frame 52 |
756 |
0.41 |
| chr1_11851036_11851187 | 0.39 |
C1orf167 |
chromosome 1 open reading frame 167 |
11237 |
0.1 |
| chr6_56820874_56821079 | 0.39 |
BEND6 |
BEN domain containing 6 |
806 |
0.54 |
| chrX_73639854_73640301 | 0.39 |
SLC16A2 |
solute carrier family 16, member 2 (thyroid hormone transporter) |
1008 |
0.55 |
| chr18_61149029_61149180 | 0.38 |
RP11-635N19.3 |
|
3730 |
0.21 |
| chr17_39656686_39656897 | 0.37 |
AC019349.5 |
|
444 |
0.64 |
| chr2_158113918_158114875 | 0.37 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
286 |
0.93 |
| chr17_21360959_21361110 | 0.37 |
KCNJ12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
52586 |
0.13 |
| chr17_70587270_70588900 | 0.37 |
ENSG00000200783 |
. |
72206 |
0.12 |
| chr3_50176033_50176184 | 0.37 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
16370 |
0.12 |
| chr11_45943886_45945188 | 0.37 |
GYLTL1B |
glycosyltransferase-like 1B |
208 |
0.9 |
| chr21_47401799_47402727 | 0.37 |
COL6A1 |
collagen, type VI, alpha 1 |
612 |
0.75 |
| chr7_44529962_44530754 | 0.37 |
NUDCD3 |
NudC domain containing 3 |
121 |
0.96 |
| chr22_29705272_29705553 | 0.37 |
GAS2L1 |
growth arrest-specific 2 like 1 |
2364 |
0.17 |
| chr18_19751318_19751469 | 0.36 |
GATA6 |
GATA binding protein 6 |
499 |
0.73 |
| chr2_112165488_112165639 | 0.36 |
ENSG00000266139 |
. |
86895 |
0.1 |
| chr17_41656365_41656516 | 0.36 |
ETV4 |
ets variant 4 |
548 |
0.74 |
| chr7_42277779_42278105 | 0.36 |
GLI3 |
GLI family zinc finger 3 |
1284 |
0.64 |
| chr17_80196270_80197118 | 0.36 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
1648 |
0.23 |
| chr3_159505128_159505279 | 0.36 |
IQCJ-SCHIP1-AS1 |
IQCJ-SCHIP1 readthrough antisense RNA 1 |
18802 |
0.15 |
| chr16_2517636_2518674 | 0.36 |
RP11-715J22.2 |
|
59 |
0.92 |
| chr8_144682749_144682900 | 0.36 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
1113 |
0.23 |
| chr19_55996251_55997143 | 0.36 |
NAT14 |
N-acetyltransferase 14 (GCN5-related, putative) |
96 |
0.92 |
| chr10_119301594_119301816 | 0.35 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
125 |
0.68 |
| chr1_156084579_156085364 | 0.35 |
LMNA |
lamin A/C |
458 |
0.7 |
| chr9_84303354_84304234 | 0.35 |
TLE1 |
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
198 |
0.96 |
| chr1_934275_935357 | 0.35 |
HES4 |
hes family bHLH transcription factor 4 |
545 |
0.57 |
| chr7_552435_552586 | 0.35 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
5635 |
0.22 |
| chr6_46702576_46703631 | 0.35 |
PLA2G7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
24 |
0.97 |
| chr14_105434639_105435137 | 0.35 |
AHNAK2 |
AHNAK nucleoprotein 2 |
9806 |
0.16 |
| chr17_38473146_38473297 | 0.35 |
RARA |
retinoic acid receptor, alpha |
1312 |
0.32 |
| chr20_62328622_62328873 | 0.35 |
TNFRSF6B |
tumor necrosis factor receptor superfamily, member 6b, decoy |
726 |
0.45 |
| chr4_7023632_7023916 | 0.35 |
TADA2B |
transcriptional adaptor 2B |
19852 |
0.09 |
| chrX_37545083_37545690 | 0.35 |
XK |
X-linked Kx blood group (McLeod syndrome) |
374 |
0.91 |
| chr14_96670852_96671050 | 0.35 |
BDKRB2 |
bradykinin receptor B2 |
65 |
0.72 |
| chr20_61468182_61468333 | 0.35 |
COL9A3 |
collagen, type IX, alpha 3 |
19094 |
0.11 |
| chr10_126812393_126812551 | 0.35 |
CTBP2 |
C-terminal binding protein 2 |
34813 |
0.2 |
| chr11_131780761_131781452 | 0.34 |
NTM |
neurotrimin |
209 |
0.95 |
| chr8_28209037_28209188 | 0.34 |
ZNF395 |
zinc finger protein 395 |
29 |
0.97 |
| chr19_56631113_56632184 | 0.34 |
ZNF787 |
zinc finger protein 787 |
971 |
0.43 |
| chr1_228337001_228337152 | 0.34 |
GJC2 |
gap junction protein, gamma 2, 47kDa |
477 |
0.67 |
| chr1_159061252_159061624 | 0.34 |
AIM2 |
absent in melanoma 2 |
14747 |
0.17 |
| chr2_183731816_183732221 | 0.34 |
FRZB |
frizzled-related protein |
128 |
0.96 |
| chr6_116772620_116772771 | 0.34 |
ENSG00000265516 |
. |
6290 |
0.14 |
| chr4_55143230_55143451 | 0.34 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
33598 |
0.22 |
| chr11_94473290_94474384 | 0.34 |
RP11-867G2.8 |
|
316 |
0.92 |
| chr2_219472134_219472377 | 0.34 |
PLCD4 |
phospholipase C, delta 4 |
233 |
0.86 |
| chr21_33784461_33785454 | 0.34 |
EVA1C |
eva-1 homolog C (C. elegans) |
36 |
0.97 |
| chr17_38255735_38256962 | 0.34 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
630 |
0.6 |
| chr19_49238400_49238597 | 0.34 |
RASIP1 |
Ras interacting protein 1 |
3777 |
0.1 |
| chr1_228135806_228136352 | 0.34 |
WNT9A |
wingless-type MMTV integration site family, member 9A |
480 |
0.79 |
| chr14_105435453_105435767 | 0.34 |
AHNAK2 |
AHNAK nucleoprotein 2 |
9084 |
0.16 |
| chr7_70049486_70049637 | 0.34 |
AUTS2 |
autism susceptibility candidate 2 |
144564 |
0.05 |
| chr2_131010330_131010897 | 0.33 |
TUBA3E |
tubulin, alpha 3e |
54579 |
0.08 |
| chr6_1624554_1625514 | 0.33 |
FOXC1 |
forkhead box C1 |
14353 |
0.29 |
| chr10_106034925_106035119 | 0.33 |
GSTO2 |
glutathione S-transferase omega 2 |
98 |
0.95 |
| chr6_1311351_1312220 | 0.33 |
FOXQ1 |
forkhead box Q1 |
890 |
0.69 |
| chr12_123429278_123429575 | 0.33 |
ABCB9 |
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
204 |
0.92 |
| chr5_134366245_134366951 | 0.33 |
PITX1 |
paired-like homeodomain 1 |
2227 |
0.27 |
| chr16_14396121_14396821 | 0.33 |
ENSG00000207639 |
. |
1353 |
0.42 |
| chr14_38052851_38053204 | 0.33 |
FOXA1 |
forkhead box A1 |
11212 |
0.23 |
| chr19_37743129_37743340 | 0.33 |
ZNF383 |
zinc finger protein 383 |
24782 |
0.13 |
| chr20_62167761_62168265 | 0.33 |
PTK6 |
protein tyrosine kinase 6 |
682 |
0.51 |
| chr18_74843262_74844680 | 0.33 |
MBP |
myelin basic protein |
331 |
0.94 |
| chr3_170407376_170407527 | 0.33 |
RP11-373E16.3 |
|
33100 |
0.21 |
| chr4_113626760_113627887 | 0.33 |
ENSG00000202536 |
. |
49750 |
0.09 |
| chr18_31802356_31802659 | 0.33 |
NOL4 |
nucleolar protein 4 |
225 |
0.71 |
| chr7_155006231_155006400 | 0.32 |
AC099552.4 |
Uncharacterized protein |
16169 |
0.2 |
| chr9_111313924_111314166 | 0.32 |
ENSG00000222512 |
. |
192836 |
0.03 |
| chr1_62784952_62785183 | 0.32 |
KANK4 |
KN motif and ankyrin repeat domains 4 |
18 |
0.98 |
| chr17_70117213_70117669 | 0.32 |
SOX9 |
SRY (sex determining region Y)-box 9 |
280 |
0.95 |
| chr8_144656729_144657016 | 0.32 |
RP11-661A12.9 |
|
1212 |
0.19 |
| chr9_72658914_72659269 | 0.32 |
MAMDC2 |
MAM domain containing 2 |
594 |
0.81 |
| chr20_17207159_17207414 | 0.32 |
PCSK2 |
proprotein convertase subtilisin/kexin type 2 |
350 |
0.89 |
| chr6_166075279_166075470 | 0.32 |
PDE10A |
phosphodiesterase 10A |
183 |
0.97 |
| chr10_57388051_57388558 | 0.32 |
PCDH15 |
protocadherin-related 15 |
602 |
0.76 |
| chr6_147828410_147828574 | 0.32 |
SAMD5 |
sterile alpha motif domain containing 5 |
1571 |
0.57 |
| chr20_9487123_9487357 | 0.32 |
LAMP5 |
lysosomal-associated membrane protein family, member 5 |
7765 |
0.23 |
| chr5_68710635_68711790 | 0.32 |
MARVELD2 |
MARVEL domain containing 2 |
3 |
0.97 |
| chr11_119598721_119599273 | 0.32 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
266 |
0.88 |
| chr22_46981461_46981612 | 0.32 |
GRAMD4 |
GRAM domain containing 4 |
8510 |
0.22 |
| chr2_227662938_227664059 | 0.32 |
IRS1 |
insulin receptor substrate 1 |
977 |
0.58 |
| chr9_21974540_21975658 | 0.32 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
2 |
0.97 |
| chr21_44795271_44795422 | 0.31 |
SIK1 |
salt-inducible kinase 1 |
51662 |
0.16 |
| chr19_3343996_3344371 | 0.31 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
15378 |
0.17 |
| chr2_241860515_241860848 | 0.31 |
AC104809.3 |
Protein LOC728763 |
1305 |
0.38 |
| chr14_25518330_25518565 | 0.31 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
241 |
0.96 |
| chr3_48468926_48469077 | 0.31 |
PLXNB1 |
plexin B1 |
1871 |
0.18 |
| chr19_4472643_4473577 | 0.31 |
HDGFRP2 |
Hepatoma-derived growth factor-related protein 2 |
807 |
0.39 |
| chr17_81067326_81067613 | 0.31 |
METRNL |
meteorin, glial cell differentiation regulator-like |
15475 |
0.24 |
| chr8_27530559_27530710 | 0.31 |
ENSG00000222862 |
. |
6715 |
0.16 |
| chr10_95229341_95229492 | 0.31 |
MYOF |
myoferlin |
12535 |
0.19 |
| chr17_74525960_74526111 | 0.31 |
CYGB |
cytoglobin |
2112 |
0.15 |
| chr15_67400456_67400607 | 0.31 |
SMAD3 |
SMAD family member 3 |
9514 |
0.25 |
| chr9_140093629_140094566 | 0.31 |
TPRN |
taperin |
460 |
0.53 |
| chr19_41120135_41120880 | 0.30 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
685 |
0.6 |
| chr16_1922185_1922470 | 0.30 |
MEIOB |
meiosis specific with OB domains |
148 |
0.88 |
| chr1_31380173_31380807 | 0.30 |
SDC3 |
syndecan 3 |
1118 |
0.44 |
| chr16_58767280_58768341 | 0.30 |
GOT2 |
glutamic-oxaloacetic transaminase 2, mitochondrial |
420 |
0.82 |
| chr2_74668159_74668310 | 0.30 |
RTKN |
rhotekin |
476 |
0.58 |
| chr11_7694729_7695430 | 0.30 |
CYB5R2 |
cytochrome b5 reductase 2 |
78 |
0.97 |
| chr12_126468546_126468697 | 0.30 |
TMEM132B |
transmembrane protein 132B |
361579 |
0.01 |
| chr1_66258526_66259233 | 0.30 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
15 |
0.99 |
| chr1_95286002_95286302 | 0.30 |
SLC44A3 |
solute carrier family 44, member 3 |
25 |
0.98 |
| chr5_6767017_6767234 | 0.30 |
PAPD7 |
PAP associated domain containing 7 |
52380 |
0.15 |
| chr10_73769586_73770005 | 0.30 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
45672 |
0.15 |
| chr22_47550419_47550650 | 0.30 |
ENSG00000221672 |
. |
306731 |
0.01 |
| chr2_1594989_1595282 | 0.30 |
AC144450.1 |
|
28750 |
0.22 |
| chr8_49292805_49293034 | 0.30 |
ENSG00000252710 |
. |
72329 |
0.13 |
| chr6_10419619_10419946 | 0.30 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
89 |
0.97 |
| chr17_7942417_7942568 | 0.30 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
18 |
0.95 |
| chr1_151283798_151284073 | 0.29 |
ENSG00000265753 |
. |
10496 |
0.09 |
| chr12_57522507_57523183 | 0.29 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
38 |
0.64 |
| chr20_49465299_49465721 | 0.29 |
ENSG00000265062 |
. |
11935 |
0.18 |
| chr17_79799803_79799954 | 0.29 |
RP11-498C9.2 |
|
1163 |
0.22 |
| chr13_37005266_37006411 | 0.29 |
CCNA1 |
cyclin A1 |
129 |
0.97 |
| chr3_188390324_188390475 | 0.29 |
ENSG00000207651 |
. |
16170 |
0.22 |
| chr20_50178684_50179923 | 0.29 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
67 |
0.99 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.2 | 0.7 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.2 | 0.5 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.2 | 0.7 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.2 | 0.3 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.4 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.3 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.1 | 0.5 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.1 | 0.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.3 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.1 | 0.4 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 0.3 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.3 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.1 | 0.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 0.5 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.1 | 0.3 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.1 | 0.3 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.1 | 0.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.1 | 0.2 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.1 | 0.3 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.1 | 0.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.3 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.1 | 0.1 | GO:0060087 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.5 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.2 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 0.2 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.1 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) layer formation in cerebral cortex(GO:0021819) telencephalon glial cell migration(GO:0022030) |
| 0.1 | 0.2 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.2 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.1 | 0.2 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.1 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.3 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.1 | 0.4 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.1 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.2 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 | 0.2 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 0.2 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.1 | 0.4 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.5 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.1 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.3 | GO:0051956 | negative regulation of amino acid transport(GO:0051956) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.3 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.4 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.8 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.2 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.0 | 0.1 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.4 | GO:0071453 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.6 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0036473 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) cell death in response to oxidative stress(GO:0036473) |
| 0.0 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0017158 | regulation of calcium ion-dependent exocytosis(GO:0017158) |
| 0.0 | 0.1 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.3 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.3 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.1 | GO:0017014 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.0 | GO:1902667 | regulation of axon extension involved in axon guidance(GO:0048841) regulation of axon guidance(GO:1902667) |
| 0.0 | 0.4 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.0 | 0.1 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0072501 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.0 | GO:0048318 | axial mesoderm development(GO:0048318) axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0001916 | positive regulation of T cell mediated cytotoxicity(GO:0001916) |
| 0.0 | 0.1 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.2 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 1.1 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0043096 | adenine salvage(GO:0006168) purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.0 | 0.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.2 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.0 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.0 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.4 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.0 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.2 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0021831 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.3 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.0 | GO:0048640 | negative regulation of developmental growth(GO:0048640) |
| 0.0 | 0.4 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.4 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.2 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.2 | GO:0050927 | positive regulation of positive chemotaxis(GO:0050927) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.1 | GO:0070528 | protein kinase C signaling(GO:0070528) |
| 0.0 | 0.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0015904 | tetracycline transport(GO:0015904) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.2 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.2 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.2 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) |
| 0.0 | 0.0 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.0 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 0.1 | GO:1900006 | positive regulation of dendrite morphogenesis(GO:0050775) positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.0 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.0 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.0 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.0 | GO:0014832 | urinary bladder smooth muscle contraction(GO:0014832) |
| 0.0 | 0.1 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0032925 | regulation of activin receptor signaling pathway(GO:0032925) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0048265 | response to pain(GO:0048265) |
| 0.0 | 0.4 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0060177 | regulation of angiotensin metabolic process(GO:0060177) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.1 | GO:0001960 | negative regulation of cytokine-mediated signaling pathway(GO:0001960) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.0 | 0.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.1 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.1 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.0 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.4 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.2 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0030681 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 1.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 3.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.4 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0019861 | obsolete flagellum(GO:0019861) |
| 0.0 | 0.1 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.0 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.4 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 0.5 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.2 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.5 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.1 | 0.8 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.6 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.1 | 0.2 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.1 | 0.4 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.1 | 1.0 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.2 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.1 | 0.2 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.2 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.2 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.1 | 0.2 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.1 | 0.2 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.3 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.3 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.3 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0031420 | alkali metal ion binding(GO:0031420) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0008493 | tetracycline transporter activity(GO:0008493) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.0 | GO:0019798 | procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.0 | GO:0016849 | phosphorus-oxygen lyase activity(GO:0016849) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.0 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.0 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.3 | GO:0008066 | glutamate receptor activity(GO:0008066) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.2 | GO:0009975 | cyclase activity(GO:0009975) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.0 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.0 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.0 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.7 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 2.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.0 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 0.3 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.0 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB2 SIGNALING | Genes involved in PI3K events in ERBB2 signaling |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.5 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.3 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.5 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.0 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME OPIOID SIGNALLING | Genes involved in Opioid Signalling |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |